Program: 12. Neuroblastoma: Adrenergic #2.
Program description and justification of annotation: 12.
Submit a comment on this gene expression program’s interpretation: CLICK
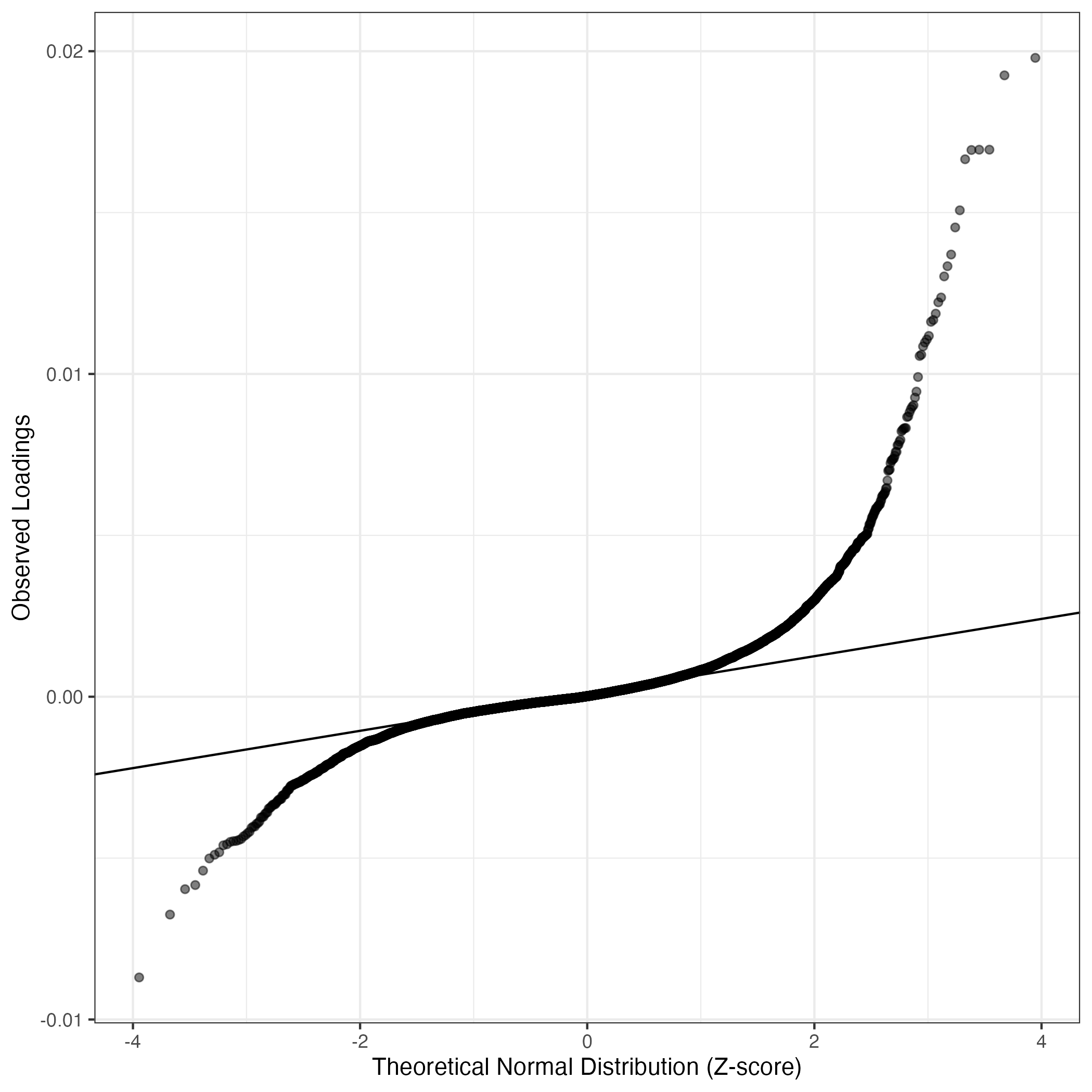
QQ-plot of gene loadings, averaged over both independent splits of the data
This plot highlights the relative contribution of each gene to the GEP
Top genes driving this program.
Note: Decartes website is buggy, try refreshing. Also, Decartes fetal adrenal data have been collected at specific time points (89-122 days), all possible cell types of interest may not be represented, do not overinterpret.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
| Gene | Loading | Gene.Name | GTEx | DepMap | Descartes | Mean.Counts | Mean.Tpm | |
|---|---|---|---|---|---|---|---|---|
| 1 | CHGA | 0.0197936 | chromogranin A | GTEx | DepMap | Descartes | 3.32 | 6803.85 |
| 2 | ERBB4 | 0.0192517 | erb-b2 receptor tyrosine kinase 4 | GTEx | DepMap | Descartes | 4.42 | 1593.77 |
| 3 | DLK1 | 0.0169486 | delta like non-canonical Notch ligand 1 | GTEx | DepMap | Descartes | 1.54 | 1490.73 |
| 4 | PDIA6 | 0.0169478 | protein disulfide isomerase family A member 6 | GTEx | DepMap | Descartes | 1.76 | 2883.91 |
| 5 | STRA6 | 0.0169388 | signaling receptor and transporter of retinol STRA6 | GTEx | DepMap | Descartes | 0.80 | 1103.42 |
| 6 | OPRM1 | 0.0166524 | opioid receptor mu 1 | GTEx | DepMap | Descartes | 0.81 | 228.32 |
| 7 | FAM155A | 0.0150706 | NA | GTEx | DepMap | Descartes | 5.30 | 2348.91 |
| 8 | RET | 0.0145396 | ret proto-oncogene | GTEx | DepMap | Descartes | 0.58 | 405.80 |
| 9 | NOL10 | 0.0137002 | nucleolar protein 10 | GTEx | DepMap | Descartes | 1.07 | 1284.86 |
| 10 | PDE10A | 0.0133388 | phosphodiesterase 10A | GTEx | DepMap | Descartes | 1.14 | 511.79 |
| 11 | ST18 | 0.0130227 | ST18 C2H2C-type zinc finger transcription factor | GTEx | DepMap | Descartes | 0.51 | 354.01 |
| 12 | TRIB2 | 0.0123688 | tribbles pseudokinase 2 | GTEx | DepMap | Descartes | 1.61 | 1502.09 |
| 13 | ADARB2 | 0.0122200 | adenosine deaminase RNA specific B2 (inactive) | GTEx | DepMap | Descartes | 0.67 | 285.33 |
| 14 | MEG3 | 0.0118689 | maternally expressed 3 | GTEx | DepMap | Descartes | 6.91 | 1982.35 |
| 15 | NR2F1 | 0.0116732 | nuclear receptor subfamily 2 group F member 1 | GTEx | DepMap | Descartes | 0.50 | 533.72 |
| 16 | RRM2 | 0.0116163 | ribonucleotide reductase regulatory subunit M2 | GTEx | DepMap | Descartes | 0.60 | 718.97 |
| 17 | CACNA1D | 0.0111783 | calcium voltage-gated channel subunit alpha1 D | GTEx | DepMap | Descartes | 0.54 | 232.76 |
| 18 | ROCK2 | 0.0110683 | Rho associated coiled-coil containing protein kinase 2 | GTEx | DepMap | Descartes | 0.96 | 461.45 |
| 19 | KCNT1 | 0.0109752 | potassium sodium-activated channel subfamily T member 1 | GTEx | DepMap | Descartes | 0.27 | 137.38 |
| 20 | ODC1 | 0.0108540 | ornithine decarboxylase 1 | GTEx | DepMap | Descartes | 0.88 | 1565.47 |
| 21 | FKBP10 | 0.0105957 | FKBP prolyl isomerase 10 | GTEx | DepMap | Descartes | 0.45 | 608.22 |
| 22 | FAM13A | 0.0105571 | family with sequence similarity 13 member A | GTEx | DepMap | Descartes | 0.51 | 362.50 |
| 23 | TLE2 | 0.0099061 | TLE family member 2, transcriptional corepressor | GTEx | DepMap | Descartes | 0.40 | 564.19 |
| 24 | PLD5 | 0.0094533 | phospholipase D family member 5 | GTEx | DepMap | Descartes | 0.71 | 308.22 |
| 25 | HPCAL1 | 0.0092682 | hippocalcin like 1 | GTEx | DepMap | Descartes | 0.66 | 514.35 |
| 26 | CACNA2D2 | 0.0090320 | calcium voltage-gated channel auxiliary subunit alpha2delta 2 | GTEx | DepMap | Descartes | 0.28 | 198.57 |
| 27 | CEP112 | 0.0089826 | centrosomal protein 112 | GTEx | DepMap | Descartes | 0.63 | 736.44 |
| 28 | ZMAT4 | 0.0089087 | zinc finger matrin-type 4 | GTEx | DepMap | Descartes | 0.44 | 718.97 |
| 29 | TAF1B | 0.0088106 | TATA-box binding protein associated factor, RNA polymerase I subunit B | GTEx | DepMap | Descartes | 0.52 | 944.29 |
| 30 | AFF3 | 0.0086867 | AF4/FMR2 family member 3 | GTEx | DepMap | Descartes | 0.85 | 345.35 |
| 31 | LHX9 | 0.0086657 | LIM homeobox 9 | GTEx | DepMap | Descartes | 0.18 | 97.70 |
| 32 | ATP8A2 | 0.0083242 | ATPase phospholipid transporting 8A2 | GTEx | DepMap | Descartes | 0.32 | 134.48 |
| 33 | NKAIN3 | 0.0083236 | sodium/potassium transporting ATPase interacting 3 | GTEx | DepMap | Descartes | 0.20 | 414.63 |
| 34 | CYB561 | 0.0083061 | cytochrome b561 | GTEx | DepMap | Descartes | 0.45 | 571.16 |
| 35 | E2F6 | 0.0082654 | E2F transcription factor 6 | GTEx | DepMap | Descartes | 0.35 | 430.02 |
| 36 | FGF14 | 0.0082252 | fibroblast growth factor 14 | GTEx | DepMap | Descartes | 2.68 | 861.47 |
| 37 | SLC35F3 | 0.0079604 | solute carrier family 35 member F3 | GTEx | DepMap | Descartes | 0.34 | 397.51 |
| 38 | RAP1GAP2 | 0.0079040 | RAP1 GTPase activating protein 2 | GTEx | DepMap | Descartes | 0.92 | 506.60 |
| 39 | GRHL1 | 0.0078089 | grainyhead like transcription factor 1 | GTEx | DepMap | Descartes | 0.27 | 203.32 |
| 40 | COL4A5 | 0.0077919 | collagen type IV alpha 5 chain | GTEx | DepMap | Descartes | 0.15 | 93.67 |
| 41 | PCSK2 | 0.0075864 | proprotein convertase subtilisin/kexin type 2 | GTEx | DepMap | Descartes | 0.41 | 351.78 |
| 42 | DSCAML1 | 0.0075858 | DS cell adhesion molecule like 1 | GTEx | DepMap | Descartes | 0.22 | 120.23 |
| 43 | CPLX2 | 0.0074677 | complexin 2 | GTEx | DepMap | Descartes | 0.38 | 309.83 |
| 44 | SEZ6 | 0.0073808 | seizure related 6 homolog | GTEx | DepMap | Descartes | 0.21 | 201.73 |
| 45 | SLC18A1 | 0.0073638 | solute carrier family 18 member A1 | GTEx | DepMap | Descartes | 0.29 | 394.49 |
| 46 | CHGB | 0.0073304 | chromogranin B | GTEx | DepMap | Descartes | 1.18 | 2045.75 |
| 47 | IGF2 | 0.0073169 | insulin like growth factor 2 | GTEx | DepMap | Descartes | 0.14 | 104.39 |
| 48 | LPIN1 | 0.0072349 | lipin 1 | GTEx | DepMap | Descartes | 1.74 | 1195.37 |
| 49 | NGFR | 0.0070383 | nerve growth factor receptor | GTEx | DepMap | Descartes | 0.17 | 216.73 |
| 50 | KCNK9 | 0.0070254 | potassium two pore domain channel subfamily K member 9 | GTEx | DepMap | Descartes | 0.14 | 105.85 |
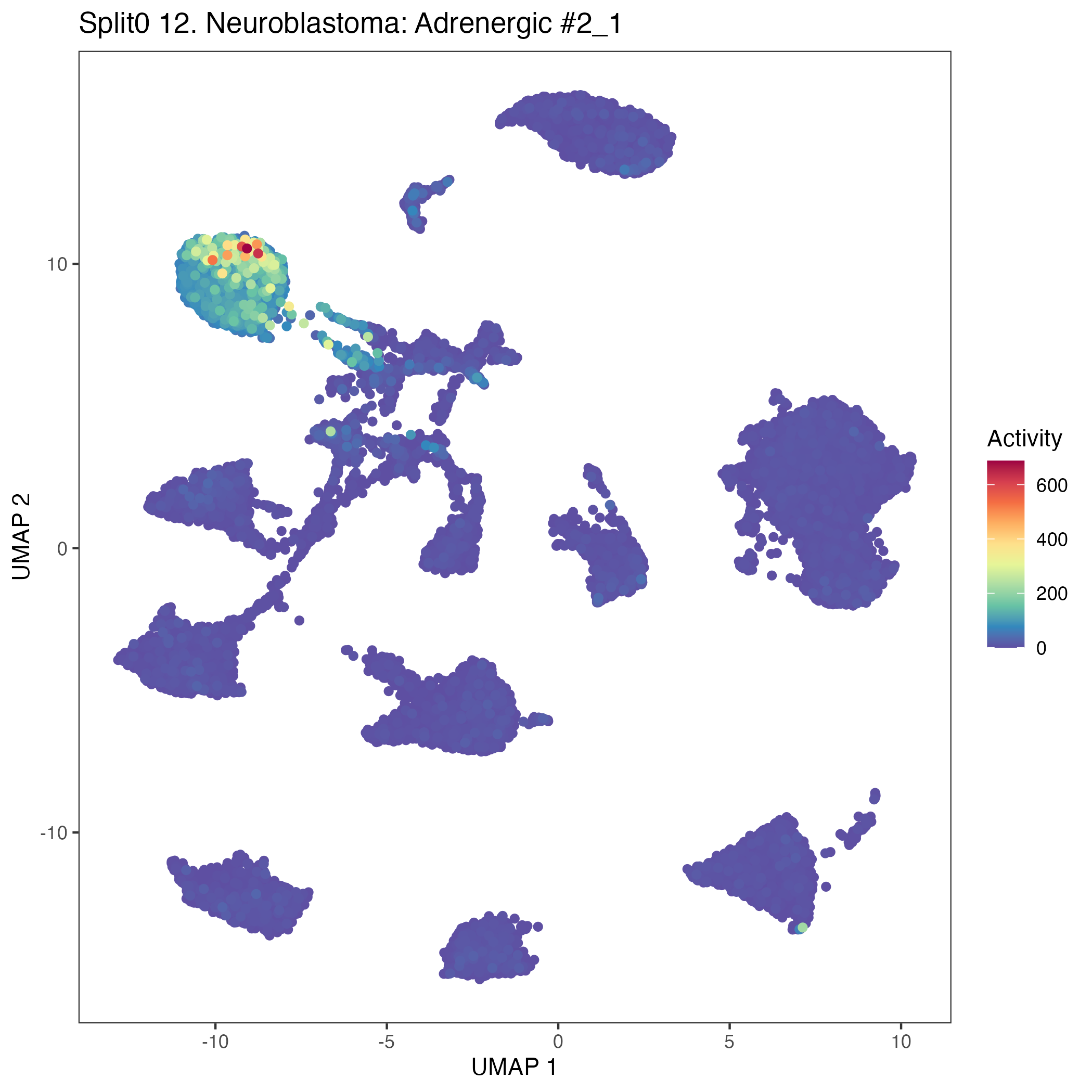
UMAP plots showing activity of gene expression program identified in community:12. Neuroblastoma: Adrenergic #2
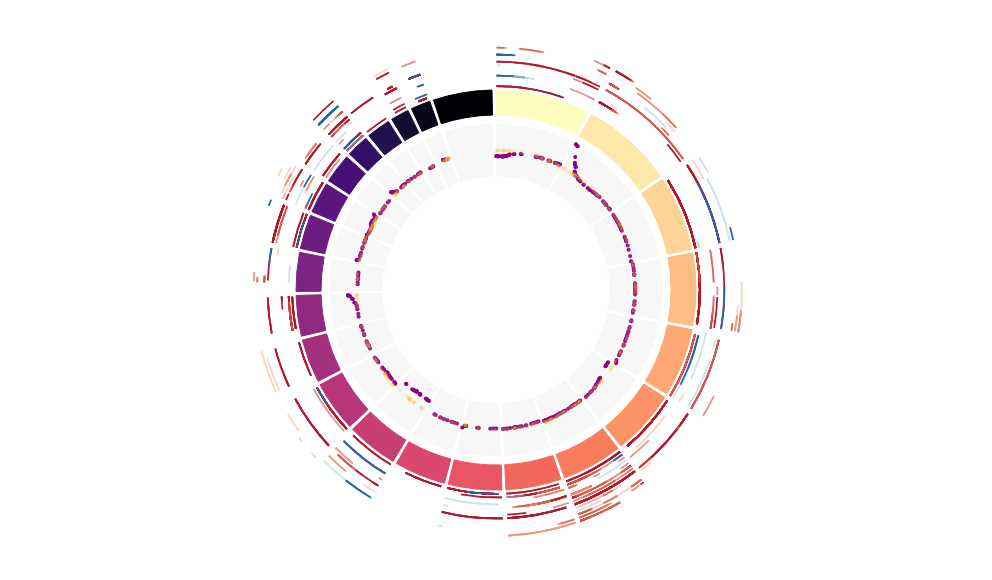
CNV Data procured from inferCNV.
Outer tracks are putative CNV regions (gains = red, losses = blue) for each patient
Inner track is expression data representing:
The top cells expressing this GEP (purple)
Random cells (n =50) from the reference set used in inferCNV (orange)
Gene set Enrichments for this program, caculated from top 50 genes
mSigDB Cell Types Gene Set:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| DESCARTES_FETAL_ADRENAL_SYMPATHOBLASTS | 3.45e-07 | 18.49 | 6.86 | 2.31e-04 | 2.31e-04 | 7CHGA, OPRM1, RET, ST18, SLC35F3, PCSK2, KCNK9 |
117 |
| BUSSLINGER_DUODENAL_EC_CELLS | 2.46e-04 | 28.45 | 5.35 | 2.07e-02 | 1.65e-01 | 3CHGA, PCSK2, CHGB |
31 |
| DESCARTES_MAIN_FETAL_SYMPATHOBLASTS | 7.41e-05 | 20.48 | 5.17 | 1.24e-02 | 4.97e-02 | 4CHGA, CYB561, SLC18A1, CHGB |
57 |
| MENON_FETAL_KIDNEY_2_NEPHRON_PROGENITOR_CELLS | 6.96e-04 | 19.44 | 3.72 | 4.24e-02 | 4.67e-01 | 3ERBB4, TRIB2, ODC1 |
44 |
| ZHONG_PFC_MAJOR_TYPES_INTERNEURON | 2.28e-03 | 32.49 | 3.53 | 8.50e-02 | 1.00e+00 | 2ERBB4, MEG3 |
18 |
| BUSSLINGER_GASTRIC_OXYNTIC_ENTEROCHROMAFFIN_LIKE_CELLS | 3.47e-04 | 13.41 | 3.43 | 2.41e-02 | 2.33e-01 | 4CHGA, FGF14, CPLX2, CHGB |
85 |
| DESCARTES_FETAL_STOMACH_NEUROENDOCRINE_CELLS | 1.01e-03 | 16.95 | 3.26 | 4.85e-02 | 6.78e-01 | 3CHGA, ST18, SLC18A1 |
50 |
| DESCARTES_FETAL_STOMACH_PDE1C_ACSM3_POSITIVE_CELLS | 3.40e-03 | 26.01 | 2.87 | 1.20e-01 | 1.00e+00 | 2ERBB4, ADARB2 |
22 |
| MANNO_MIDBRAIN_NEUROTYPES_HDA2 | 1.99e-05 | 6.22 | 2.76 | 6.69e-03 | 1.34e-02 | 10ERBB4, DLK1, FAM155A, RET, ADARB2, MEG3, ATP8A2, RAP1GAP2, PCSK2, SLC18A1 |
513 |
| DESCARTES_FETAL_INTESTINE_CHROMAFFIN_CELLS | 9.77e-04 | 10.06 | 2.58 | 4.85e-02 | 6.56e-01 | 4CHGA, ST18, RAP1GAP2, SLC18A1 |
112 |
| GAO_LARGE_INTESTINE_ADULT_CA_ENTEROENDOCRINE_CELLS | 1.89e-04 | 6.67 | 2.51 | 2.07e-02 | 1.27e-01 | 7ST18, CACNA1D, AFF3, CPLX2, SEZ6, SLC18A1, CHGB |
312 |
| MANNO_MIDBRAIN_NEUROTYPES_HDA1 | 5.75e-05 | 5.45 | 2.42 | 1.24e-02 | 3.86e-02 | 10CHGA, ERBB4, DLK1, FAM155A, ADARB2, MEG3, ATP8A2, PCSK2, SLC18A1, LPIN1 |
584 |
| MANNO_MIDBRAIN_NEUROTYPES_HDA | 1.11e-04 | 5.52 | 2.35 | 1.50e-02 | 7.48e-02 | 9CHGA, ERBB4, DLK1, FAM155A, ADARB2, MEG3, TLE2, RAP1GAP2, SLC18A1 |
506 |
| TRAVAGLINI_LUNG_NEUROENDOCRINE_CELL | 1.59e-03 | 8.76 | 2.26 | 6.29e-02 | 1.00e+00 | 4MEG3, PCSK2, CPLX2, CHGB |
128 |
| BUSSLINGER_GASTRIC_D_CELLS | 5.48e-03 | 20.00 | 2.24 | 1.60e-01 | 1.00e+00 | 2PCSK2, CHGB |
28 |
| MANNO_MIDBRAIN_NEUROTYPES_HNBML5 | 3.58e-04 | 5.21 | 2.10 | 2.41e-02 | 2.41e-01 | 8CHGA, FAM155A, ST18, ADARB2, MEG3, FGF14, RAP1GAP2, LPIN1 |
465 |
| DESCARTES_FETAL_LUNG_VISCERAL_NEURONS | 1.39e-03 | 6.71 | 2.06 | 6.23e-02 | 9.35e-01 | 5PLD5, NKAIN3, SEZ6, NGFR, KCNK9 |
212 |
| MANNO_MIDBRAIN_NEUROTYPES_HNBM | 9.48e-04 | 5.90 | 2.04 | 4.85e-02 | 6.36e-01 | 6ST18, ZMAT4, RAP1GAP2, PCSK2, DSCAML1, SEZ6 |
295 |
| MANNO_MIDBRAIN_NEUROTYPES_HNBGABA | 2.46e-04 | 4.51 | 2.00 | 2.07e-02 | 1.65e-01 | 10CHGA, OPRM1, FAM155A, MEG3, ATP8A2, FGF14, RAP1GAP2, PCSK2, SEZ6, LPIN1 |
703 |
| MENON_FETAL_KIDNEY_3_STROMAL_CELLS | 4.01e-03 | 10.22 | 1.99 | 1.35e-01 | 1.00e+00 | 3MEG3, NR2F1, IGF2 |
81 |
Dowload full table
mSigDB Hallmark Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| HALLMARK_PANCREAS_BETA_CELLS | 1.10e-02 | 13.70 | 1.56 | 5.48e-01 | 5.48e-01 | 2CHGA, PCSK2 |
40 |
| HALLMARK_ESTROGEN_RESPONSE_LATE | 1.85e-01 | 2.63 | 0.31 | 1.00e+00 | 1.00e+00 | 2RET, CACNA2D2 |
200 |
| HALLMARK_KRAS_SIGNALING_UP | 1.85e-01 | 2.63 | 0.31 | 1.00e+00 | 1.00e+00 | 2TRIB2, IGF2 |
200 |
| HALLMARK_UNFOLDED_PROTEIN_RESPONSE | 3.62e-01 | 2.28 | 0.06 | 1.00e+00 | 1.00e+00 | 1PDIA6 |
113 |
| HALLMARK_SPERMATOGENESIS | 4.15e-01 | 1.90 | 0.05 | 1.00e+00 | 1.00e+00 | 1LPIN1 |
135 |
| HALLMARK_FATTY_ACID_METABOLISM | 4.66e-01 | 1.62 | 0.04 | 1.00e+00 | 1.00e+00 | 1ODC1 |
158 |
| HALLMARK_UV_RESPONSE_UP | 4.66e-01 | 1.62 | 0.04 | 1.00e+00 | 1.00e+00 | 1RET |
158 |
| HALLMARK_IL2_STAT5_SIGNALING | 5.45e-01 | 1.29 | 0.03 | 1.00e+00 | 1.00e+00 | 1ODC1 |
199 |
| HALLMARK_G2M_CHECKPOINT | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1ODC1 |
200 |
| HALLMARK_ESTROGEN_RESPONSE_EARLY | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1RET |
200 |
| HALLMARK_MYOGENESIS | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1LPIN1 |
200 |
| HALLMARK_MTORC1_SIGNALING | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1RRM2 |
200 |
| HALLMARK_E2F_TARGETS | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1RRM2 |
200 |
| HALLMARK_MYC_TARGETS_V1 | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1ODC1 |
200 |
| HALLMARK_GLYCOLYSIS | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1LHX9 |
200 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
200 |
| HALLMARK_HYPOXIA | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
200 |
| HALLMARK_CHOLESTEROL_HOMEOSTASIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
74 |
| HALLMARK_MITOTIC_SPINDLE | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
199 |
| HALLMARK_WNT_BETA_CATENIN_SIGNALING | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
42 |
Dowload full table
KEGG Pathways:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| KEGG_GLUTATHIONE_METABOLISM | 1.67e-02 | 10.84 | 1.24 | 1.00e+00 | 1.00e+00 | 2RRM2, ODC1 |
50 |
| KEGG_ARRHYTHMOGENIC_RIGHT_VENTRICULAR_CARDIOMYOPATHY_ARVC | 3.47e-02 | 7.23 | 0.84 | 1.00e+00 | 1.00e+00 | 2CACNA1D, CACNA2D2 |
74 |
| KEGG_CARDIAC_MUSCLE_CONTRACTION | 3.90e-02 | 6.76 | 0.78 | 1.00e+00 | 1.00e+00 | 2CACNA1D, CACNA2D2 |
79 |
| KEGG_HYPERTROPHIC_CARDIOMYOPATHY_HCM | 4.26e-02 | 6.43 | 0.74 | 1.00e+00 | 1.00e+00 | 2CACNA1D, CACNA2D2 |
83 |
| KEGG_DILATED_CARDIOMYOPATHY | 4.93e-02 | 5.92 | 0.69 | 1.00e+00 | 1.00e+00 | 2CACNA1D, CACNA2D2 |
90 |
| KEGG_MAPK_SIGNALING_PATHWAY | 8.58e-02 | 3.02 | 0.60 | 1.00e+00 | 1.00e+00 | 3CACNA1D, CACNA2D2, FGF14 |
267 |
| KEGG_VASCULAR_SMOOTH_MUSCLE_CONTRACTION | 7.56e-02 | 4.61 | 0.54 | 1.00e+00 | 1.00e+00 | 2CACNA1D, ROCK2 |
115 |
| KEGG_PURINE_METABOLISM | 1.29e-01 | 3.32 | 0.39 | 1.00e+00 | 1.00e+00 | 2PDE10A, RRM2 |
159 |
| KEGG_CALCIUM_SIGNALING_PATHWAY | 1.55e-01 | 2.96 | 0.35 | 1.00e+00 | 1.00e+00 | 2ERBB4, CACNA1D |
178 |
| KEGG_ENDOCYTOSIS | 1.59e-01 | 2.91 | 0.34 | 1.00e+00 | 1.00e+00 | 2ERBB4, RET |
181 |
| KEGG_REGULATION_OF_ACTIN_CYTOSKELETON | 2.03e-01 | 2.47 | 0.29 | 1.00e+00 | 1.00e+00 | 2ROCK2, FGF14 |
213 |
| KEGG_THYROID_CANCER | 1.09e-01 | 9.11 | 0.22 | 1.00e+00 | 1.00e+00 | 1RET |
29 |
| KEGG_PATHWAYS_IN_CANCER | 3.61e-01 | 1.61 | 0.19 | 1.00e+00 | 1.00e+00 | 2RET, FGF14 |
325 |
| KEGG_TYPE_II_DIABETES_MELLITUS | 1.71e-01 | 5.54 | 0.13 | 1.00e+00 | 1.00e+00 | 1CACNA1D |
47 |
| KEGG_ARGININE_AND_PROLINE_METABOLISM | 1.94e-01 | 4.81 | 0.12 | 1.00e+00 | 1.00e+00 | 1ODC1 |
54 |
| KEGG_PATHOGENIC_ESCHERICHIA_COLI_INFECTION | 2.00e-01 | 4.64 | 0.11 | 1.00e+00 | 1.00e+00 | 1ROCK2 |
56 |
| KEGG_P53_SIGNALING_PATHWAY | 2.37e-01 | 3.81 | 0.09 | 1.00e+00 | 1.00e+00 | 1RRM2 |
68 |
| KEGG_MELANOMA | 2.46e-01 | 3.64 | 0.09 | 1.00e+00 | 1.00e+00 | 1FGF14 |
71 |
| KEGG_TGF_BETA_SIGNALING_PATHWAY | 2.90e-01 | 3.00 | 0.07 | 1.00e+00 | 1.00e+00 | 1ROCK2 |
86 |
| KEGG_ERBB_SIGNALING_PATHWAY | 2.93e-01 | 2.97 | 0.07 | 1.00e+00 | 1.00e+00 | 1ERBB4 |
87 |
Dowload full table
CHR Positional Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| chr2p25 | 2.31e-11 | 29.19 | 12.68 | 6.41e-09 | 6.41e-09 | 10PDIA6, NOL10, RRM2, ROCK2, ODC1, HPCAL1, TAF1B, E2F6, GRHL1, LPIN1 |
117 |
| chr13q33 | 2.00e-02 | 9.82 | 1.13 | 1.00e+00 | 1.00e+00 | 2FAM155A, FGF14 |
55 |
| chr20p12 | 6.35e-02 | 5.11 | 0.59 | 1.00e+00 | 1.00e+00 | 2PCSK2, CHGB |
104 |
| chr14q32 | 4.65e-01 | 1.47 | 0.29 | 1.00e+00 | 1.00e+00 | 3CHGA, DLK1, MEG3 |
546 |
| chr3p21 | 3.32e-01 | 1.72 | 0.20 | 1.00e+00 | 1.00e+00 | 2CACNA1D, CACNA2D2 |
304 |
| chr2q34 | 1.64e-01 | 5.80 | 0.14 | 1.00e+00 | 1.00e+00 | 1ERBB4 |
45 |
| chr17q21 | 6.95e-01 | 1.14 | 0.13 | 1.00e+00 | 1.00e+00 | 2FKBP10, NGFR |
457 |
| chr1q43 | 2.13e-01 | 4.32 | 0.11 | 1.00e+00 | 1.00e+00 | 1PLD5 |
60 |
| chr5q15 | 2.13e-01 | 4.32 | 0.11 | 1.00e+00 | 1.00e+00 | 1NR2F1 |
60 |
| chr4q22 | 2.43e-01 | 3.70 | 0.09 | 1.00e+00 | 1.00e+00 | 1FAM13A |
70 |
| chr8q11 | 2.43e-01 | 3.70 | 0.09 | 1.00e+00 | 1.00e+00 | 1ST18 |
70 |
| chr1q31 | 2.46e-01 | 3.64 | 0.09 | 1.00e+00 | 1.00e+00 | 1LHX9 |
71 |
| chr2p24 | 2.55e-01 | 3.49 | 0.09 | 1.00e+00 | 1.00e+00 | 1TRIB2 |
74 |
| chr6q27 | 2.58e-01 | 3.45 | 0.08 | 1.00e+00 | 1.00e+00 | 1PDE10A |
75 |
| chr10p15 | 2.90e-01 | 3.00 | 0.07 | 1.00e+00 | 1.00e+00 | 1ADARB2 |
86 |
| chr8q12 | 2.95e-01 | 2.93 | 0.07 | 1.00e+00 | 1.00e+00 | 1NKAIN3 |
88 |
| chr17q24 | 3.12e-01 | 2.74 | 0.07 | 1.00e+00 | 1.00e+00 | 1CEP112 |
94 |
| chr8p11 | 3.15e-01 | 2.71 | 0.07 | 1.00e+00 | 1.00e+00 | 1ZMAT4 |
95 |
| chr17q23 | 3.59e-01 | 2.30 | 0.06 | 1.00e+00 | 1.00e+00 | 1CYB561 |
112 |
| chr15q24 | 3.69e-01 | 2.22 | 0.05 | 1.00e+00 | 1.00e+00 | 1STRA6 |
116 |
Dowload full table
Transcription Factor Targets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| TAXCREB_02 | 5.10e-03 | 20.80 | 2.32 | 6.76e-01 | 1.00e+00 | 2DLK1, NOL10 |
27 |
| CAGNYGKNAAA_UNKNOWN | 3.61e-03 | 10.64 | 2.07 | 5.84e-01 | 1.00e+00 | 3TRIB2, CACNA2D2, AFF3 |
78 |
| TAATTA_CHX10_01 | 1.83e-04 | 4.34 | 2.00 | 2.07e-01 | 2.07e-01 | 11ERBB4, OPRM1, ADARB2, NR2F1, HPCAL1, AFF3, GRHL1, COL4A5, PCSK2, DSCAML1, SEZ6 |
823 |
| NRSF_01 | 6.61e-03 | 8.48 | 1.66 | 7.49e-01 | 1.00e+00 | 3CHGA, PCSK2, SEZ6 |
97 |
| OCT1_02 | 9.69e-03 | 5.18 | 1.34 | 8.21e-01 | 1.00e+00 | 4ERBB4, CACNA1D, AFF3, PCSK2 |
214 |
| MYOGNF1_01 | 1.55e-02 | 11.31 | 1.29 | 8.21e-01 | 1.00e+00 | 2ROCK2, SEZ6 |
48 |
| FOXF2_TARGET_GENES | 1.98e-02 | 63.54 | 1.27 | 8.21e-01 | 1.00e+00 | 1HPCAL1 |
5 |
| AACTTT_UNKNOWN | 5.37e-03 | 2.54 | 1.26 | 6.76e-01 | 1.00e+00 | 14ERBB4, DLK1, ST18, NR2F1, CACNA1D, ROCK2, HPCAL1, ZMAT4, AFF3, ATP8A2, NKAIN3, FGF14, COL4A5, DSCAML1 |
1928 |
| IGLV5_37_TARGET_GENES | 9.66e-03 | 3.24 | 1.22 | 8.21e-01 | 1.00e+00 | 7TRIB2, CACNA1D, KCNT1, CYB561, E2F6, IGF2, LPIN1 |
636 |
| PTF1BETA_Q6 | 1.54e-02 | 4.49 | 1.17 | 8.21e-01 | 1.00e+00 | 4PDE10A, ST18, AFF3, FGF14 |
246 |
| AAAYRNCTG_UNKNOWN | 1.46e-02 | 3.75 | 1.16 | 8.21e-01 | 1.00e+00 | 5ERBB4, NR2F1, LHX9, COL4A5, DSCAML1 |
375 |
| CP2_02 | 1.69e-02 | 4.37 | 1.13 | 8.21e-01 | 1.00e+00 | 4NR2F1, ODC1, HPCAL1, CPLX2 |
253 |
| E47_02 | 1.71e-02 | 4.35 | 1.13 | 8.21e-01 | 1.00e+00 | 4TRIB2, AFF3, DSCAML1, NGFR |
254 |
| CTTTGA_LEF1_Q2 | 1.27e-02 | 2.53 | 1.12 | 8.21e-01 | 1.00e+00 | 10CHGA, ERBB4, STRA6, RET, NR2F1, ROCK2, CACNA2D2, ZMAT4, AFF3, FGF14 |
1247 |
| HSF2_01 | 1.82e-02 | 4.26 | 1.11 | 8.21e-01 | 1.00e+00 | 4NR2F1, CACNA1D, CACNA2D2, KCNK9 |
259 |
| CEBPB_01 | 2.00e-02 | 4.13 | 1.07 | 8.21e-01 | 1.00e+00 | 4ERBB4, ST18, AFF3, FGF14 |
267 |
| LEF1_Q6 | 2.00e-02 | 4.13 | 1.07 | 8.21e-01 | 1.00e+00 | 4ZMAT4, AFF3, LHX9, SEZ6 |
267 |
| AFP1_Q6 | 2.03e-02 | 4.12 | 1.07 | 8.21e-01 | 1.00e+00 | 4NR2F1, AFF3, PCSK2, DSCAML1 |
268 |
| EGR_Q6 | 2.33e-02 | 3.94 | 1.02 | 9.11e-01 | 1.00e+00 | 4PDE10A, CACNA1D, PLD5, PCSK2 |
280 |
| ISL1_TARGET_GENES | 2.90e-02 | 2.56 | 0.97 | 9.25e-01 | 1.00e+00 | 7FAM155A, CACNA1D, FAM13A, CEP112, ZMAT4, CYB561, CHGB |
803 |
Dowload full table
GO Biological Processes:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GOBP_RESPONSE_TO_AUDITORY_STIMULUS | 3.72e-03 | 24.78 | 2.74 | 1.00e+00 | 1.00e+00 | 2STRA6, ATP8A2 |
23 |
| GOBP_POSITIVE_REGULATION_OF_MULTICELLULAR_ORGANISM_GROWTH | 5.87e-03 | 19.27 | 2.16 | 1.00e+00 | 1.00e+00 | 2ATP8A2, IGF2 |
29 |
| GOBP_EATING_BEHAVIOR | 6.27e-03 | 18.58 | 2.09 | 1.00e+00 | 1.00e+00 | 2OPRM1, ATP8A2 |
30 |
| GOBP_RESPONSE_TO_ACETYLCHOLINE | 6.27e-03 | 18.58 | 2.09 | 1.00e+00 | 1.00e+00 | 2OPRM1, ROCK2 |
30 |
| GOBP_REGULATION_OF_TRANSCRIPTION_INVOLVED_IN_G1_S_TRANSITION_OF_MITOTIC_CELL_CYCLE | 8.47e-03 | 15.77 | 1.78 | 1.00e+00 | 1.00e+00 | 2RRM2, E2F6 |
35 |
| GOBP_FEEDING_BEHAVIOR | 5.88e-03 | 8.86 | 1.73 | 1.00e+00 | 1.00e+00 | 3STRA6, OPRM1, ATP8A2 |
93 |
| GOBP_ARTERY_DEVELOPMENT | 6.24e-03 | 8.67 | 1.70 | 1.00e+00 | 1.00e+00 | 3STRA6, FKBP10, NGFR |
95 |
| GOBP_VITAMIN_TRANSPORT | 1.15e-02 | 13.35 | 1.52 | 1.00e+00 | 1.00e+00 | 2STRA6, SLC35F3 |
41 |
| GOBP_SEMI_LUNAR_VALVE_DEVELOPMENT | 1.15e-02 | 13.35 | 1.52 | 1.00e+00 | 1.00e+00 | 2STRA6, ROCK2 |
41 |
| GOBP_REGULATION_OF_KERATINOCYTE_DIFFERENTIATION | 1.20e-02 | 13.01 | 1.48 | 1.00e+00 | 1.00e+00 | 2ROCK2, GRHL1 |
42 |
| GOBP_CALCIUM_ION_TRANSMEMBRANE_TRANSPORT | 6.36e-03 | 4.65 | 1.43 | 1.00e+00 | 1.00e+00 | 5OPRM1, FAM155A, CACNA1D, CACNA2D2, FGF14 |
304 |
| GOBP_EMBRYONIC_MORPHOGENESIS | 5.50e-03 | 3.62 | 1.37 | 1.00e+00 | 1.00e+00 | 7STRA6, RET, ROCK2, AFF3, ATP8A2, DSCAML1, IGF2 |
569 |
| GOBP_MAST_CELL_ACTIVATION_INVOLVED_IN_IMMUNE_RESPONSE | 1.49e-02 | 11.57 | 1.32 | 1.00e+00 | 1.00e+00 | 2CHGA, CPLX2 |
47 |
| GOBP_G0_TO_G1_TRANSITION | 1.49e-02 | 11.57 | 1.32 | 1.00e+00 | 1.00e+00 | 2RRM2, E2F6 |
47 |
| GOBP_PUTRESCINE_BIOSYNTHETIC_PROCESS | 1.98e-02 | 63.54 | 1.27 | 1.00e+00 | 1.00e+00 | 1ODC1 |
5 |
| GOBP_POSITIVE_REGULATION_OF_NEURON_MATURATION | 1.98e-02 | 63.54 | 1.27 | 1.00e+00 | 1.00e+00 | 1RET |
5 |
| GOBP_EXOCRINE_PANCREAS_DEVELOPMENT | 1.98e-02 | 63.54 | 1.27 | 1.00e+00 | 1.00e+00 | 1IGF2 |
5 |
| GOBP_NEGATIVE_REGULATION_OF_MYOSIN_LIGHT_CHAIN_PHOSPHATASE_ACTIVITY | 1.98e-02 | 63.54 | 1.27 | 1.00e+00 | 1.00e+00 | 1ROCK2 |
5 |
| GOBP_POSITIVE_REGULATION_OF_ODONTOGENESIS | 1.98e-02 | 63.54 | 1.27 | 1.00e+00 | 1.00e+00 | 1NGFR |
5 |
| GOBP_CGMP_CATABOLIC_PROCESS | 1.98e-02 | 63.54 | 1.27 | 1.00e+00 | 1.00e+00 | 1PDE10A |
5 |
Dowload full table
Immunological Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GSE8678_IL7R_LOW_VS_HIGH_EFF_CD8_TCELL_UP | 1.06e-03 | 7.16 | 2.19 | 1.00e+00 | 1.00e+00 | 5CACNA1D, KCNT1, FKBP10, RAP1GAP2, LPIN1 |
199 |
| GSE20366_CD103_POS_VS_CD103_KLRG1_DP_TREG_DN | 1.08e-03 | 7.12 | 2.18 | 1.00e+00 | 1.00e+00 | 5DLK1, CEP112, ZMAT4, CYB561, RAP1GAP2 |
200 |
| GSE339_CD8POS_VS_CD4CD8DN_DC_IN_CULTURE_UP | 1.08e-03 | 7.12 | 2.18 | 1.00e+00 | 1.00e+00 | 5ERBB4, RRM2, ODC1, HPCAL1, SEZ6 |
200 |
| GSE43956_WT_VS_SGK1_KO_TH17_DIFFERENTIATED_CD4_TCELL_UP | 1.08e-03 | 7.12 | 2.18 | 1.00e+00 | 1.00e+00 | 5MEG3, TAF1B, LHX9, CYB561, CHGB |
200 |
| GSE29614_CTRL_VS_TIV_FLU_VACCINE_PBMC_2007_UP | 6.35e-03 | 5.87 | 1.52 | 1.00e+00 | 1.00e+00 | 4ERBB4, ADARB2, COL4A5, KCNK9 |
189 |
| GSE29164_CD8_TCELL_VS_CD8_TCELL_AND_IL12_TREATED_MELANOMA_DAY7_DN | 7.07e-03 | 5.69 | 1.47 | 1.00e+00 | 1.00e+00 | 4RRM2, FAM13A, E2F6, FGF14 |
195 |
| GSE19401_PLN_VS_PEYERS_PATCH_FOLLICULAR_DC_UP | 7.20e-03 | 5.66 | 1.47 | 1.00e+00 | 1.00e+00 | 4PDIA6, TRIB2, MEG3, NR2F1 |
196 |
| GSE36476_CTRL_VS_TSST_ACT_72H_MEMORY_CD4_TCELL_OLD_UP | 7.32e-03 | 5.63 | 1.46 | 1.00e+00 | 1.00e+00 | 4DLK1, FAM13A, CYB561, RAP1GAP2 |
197 |
| GSE24814_STAT5_KO_VS_WT_PRE_BCELL_UP | 7.32e-03 | 5.63 | 1.46 | 1.00e+00 | 1.00e+00 | 4CHGA, CACNA1D, TLE2, PCSK2 |
197 |
| GSE360_L_MAJOR_VS_T_GONDII_DC_DN | 7.45e-03 | 5.60 | 1.45 | 1.00e+00 | 1.00e+00 | 4CHGA, CACNA1D, TAF1B, COL4A5 |
198 |
| GSE36527_CD62L_HIGH_VS_CD62L_LOW_TREG_CD69_NEG_KLRG1_NEG_DN | 7.58e-03 | 5.57 | 1.44 | 1.00e+00 | 1.00e+00 | 4STRA6, ADARB2, CEP112, CPLX2 |
199 |
| GSE40184_HEALTHY_VS_HCV_INFECTED_DONOR_PBMC_UP | 7.58e-03 | 5.57 | 1.44 | 1.00e+00 | 1.00e+00 | 4HPCAL1, AFF3, GRHL1, CPLX2 |
199 |
| GSE41978_ID2_KO_AND_BIM_KO_VS_BIM_KO_KLRG1_LOW_EFFECTOR_CD8_TCELL_DN | 7.58e-03 | 5.57 | 1.44 | 1.00e+00 | 1.00e+00 | 4RRM2, ODC1, COL4A5, SLC18A1 |
199 |
| GSE7852_LN_VS_FAT_TCONV_DN | 7.71e-03 | 5.55 | 1.44 | 1.00e+00 | 1.00e+00 | 4ODC1, CYB561, RAP1GAP2, IGF2 |
200 |
| GSE9650_GP33_VS_GP276_LCMV_SPECIFIC_EXHAUSTED_CD8_TCELL_UP | 7.71e-03 | 5.55 | 1.44 | 1.00e+00 | 1.00e+00 | 4STRA6, FKBP10, CPLX2, IGF2 |
200 |
| GSE16450_CTRL_VS_IFNA_6H_STIM_IMMATURE_NEURON_CELL_LINE_UP | 7.71e-03 | 5.55 | 1.44 | 1.00e+00 | 1.00e+00 | 4AFF3, ATP8A2, CYB561, GRHL1 |
200 |
| GSE13522_CTRL_VS_T_CRUZI_BRAZIL_STRAIN_INF_SKIN_UP | 2.00e-02 | 5.54 | 1.09 | 1.00e+00 | 1.00e+00 | 3CYB561, IGF2, KCNK9 |
147 |
| GSE5589_WT_VS_IL10_KO_LPS_STIM_MACROPHAGE_180MIN_DN | 2.30e-02 | 5.25 | 1.03 | 1.00e+00 | 1.00e+00 | 3CHGA, HPCAL1, GRHL1 |
155 |
| GSE5589_WT_VS_IL10_KO_LPS_AND_IL10_STIM_MACROPHAGE_180MIN_UP | 2.30e-02 | 5.25 | 1.03 | 1.00e+00 | 1.00e+00 | 3STRA6, TAF1B, E2F6 |
155 |
| GSE6269_E_COLI_VS_STREP_PNEUMO_INF_PBMC_UP | 2.82e-02 | 4.83 | 0.95 | 1.00e+00 | 1.00e+00 | 3MEG3, FKBP10, ATP8A2 |
168 |
Top Ranked Transcription Factors for this Gene Expression Program:
| Gene Symbol | TF Rank | DNA Binding Domain | Motif Status | IUPAC PWM | GTEx | DepMap | Decartes |
|---|---|---|---|---|---|---|---|
| ST18 | 11 | Yes | Inferred motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| NR2F1 | 15 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| TLE2 | 23 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| ZMAT4 | 28 | Yes | Likely to be sequence specific TF | Monomer or homomultimer | No motif | None | Possible RBP. |
| TAF1B | 29 | No | Unlikely to be sequence specific TF | Low specificity DNA-binding protein | No motif | None | Pol I TAF that contacts the rDNA promoter in conjunction with TBP and UBF (PMID: 7491500). No clear evidence that it directly confers sequence specificity. |
| AFF3 | 30 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | (PMID: 8555498) only says that In vitro-translated LAF-4 was able to bind strongly to double-stranded DNA cellulose - thats more consistent with nonspecific DNA binding. The key issue is whether the Transcription factor AF4/FMR2 (IPR007797) domain really is sequence specific. Most of the work has been done on the one Drosophila ortholog, lilliuputian. I see no evidence of DNA binding. Recent work on this family in human indicate that it functions in coactivation (PMID: 27899651; PMID: 261712 |
| LHX9 | 31 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | Has a putative AT-hook | None |
| E2F6 | 35 | Yes | Known motif | Monomer or homomultimer | In vivo/Misc source | None | None |
| GRHL1 | 39 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| EBF3 | 76 | Yes | Known motif | Monomer or homomultimer | In vivo/Misc source | Only known motifs are from Transfac or HocoMoco - origin is uncertain | None |
| AMH | 84 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| PLXNB2 | 86 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| NCOR2 | 99 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Contains 2 SANT domains, and no other putative DNA-binding domains |
| GPR155 | 103 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| RXRA | 108 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| CDK5RAP3 | 111 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| HOXD10 | 115 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| ADAMTS19 | 132 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | Has a putative AT-hook | None |
| FLNA | 134 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Go annotation is based on (PMID: 15684392), which shows that it binds FOXC1 and PBX1 but doesnt show evidence for binding to DNA |
| TLE1 | 136 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Transcriptional co-factor |
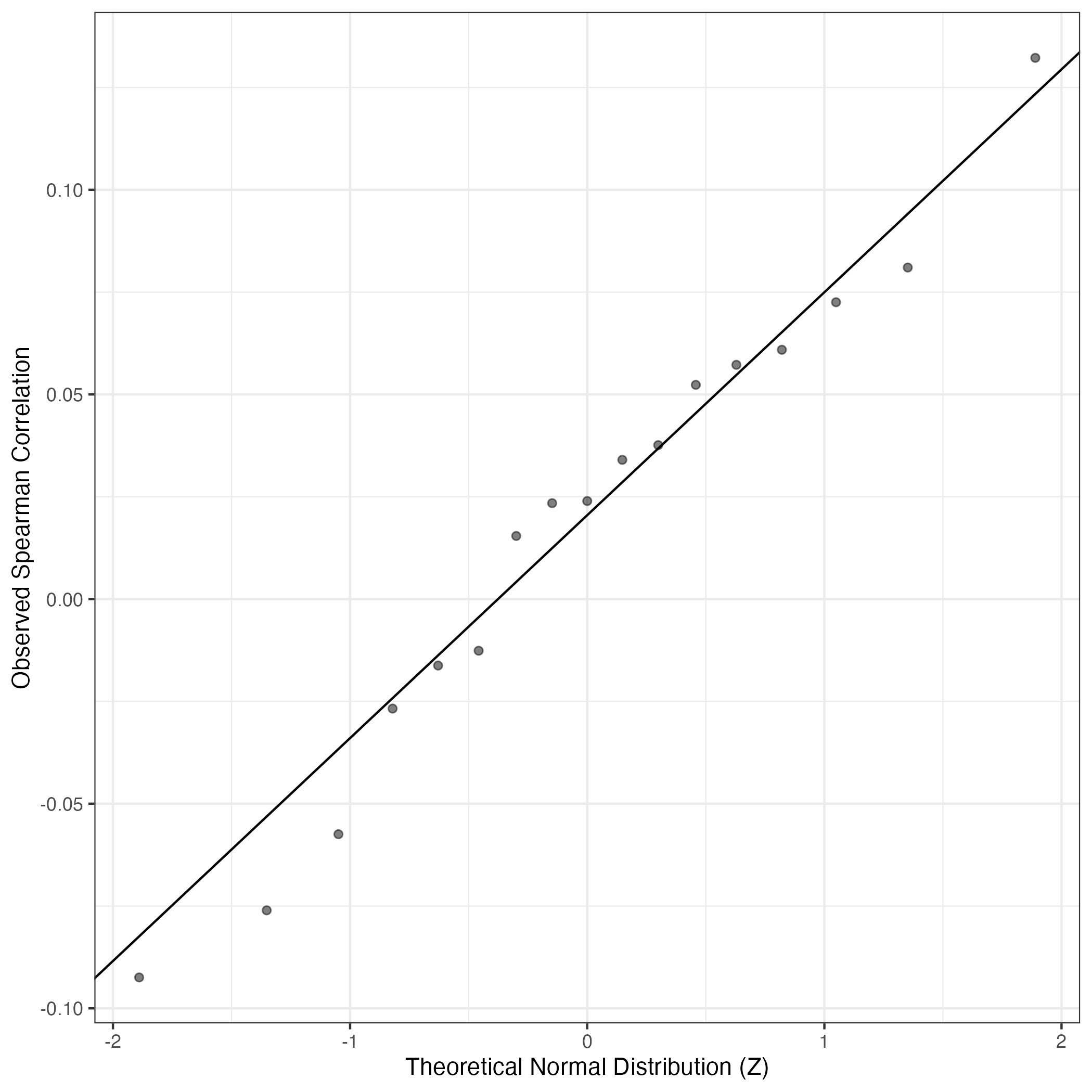
QQ Plot showing correlations with other Gene Expression Programs in this dataset, calculated by Spearman correlation:
Interactive QQ-plot of gene loadings:
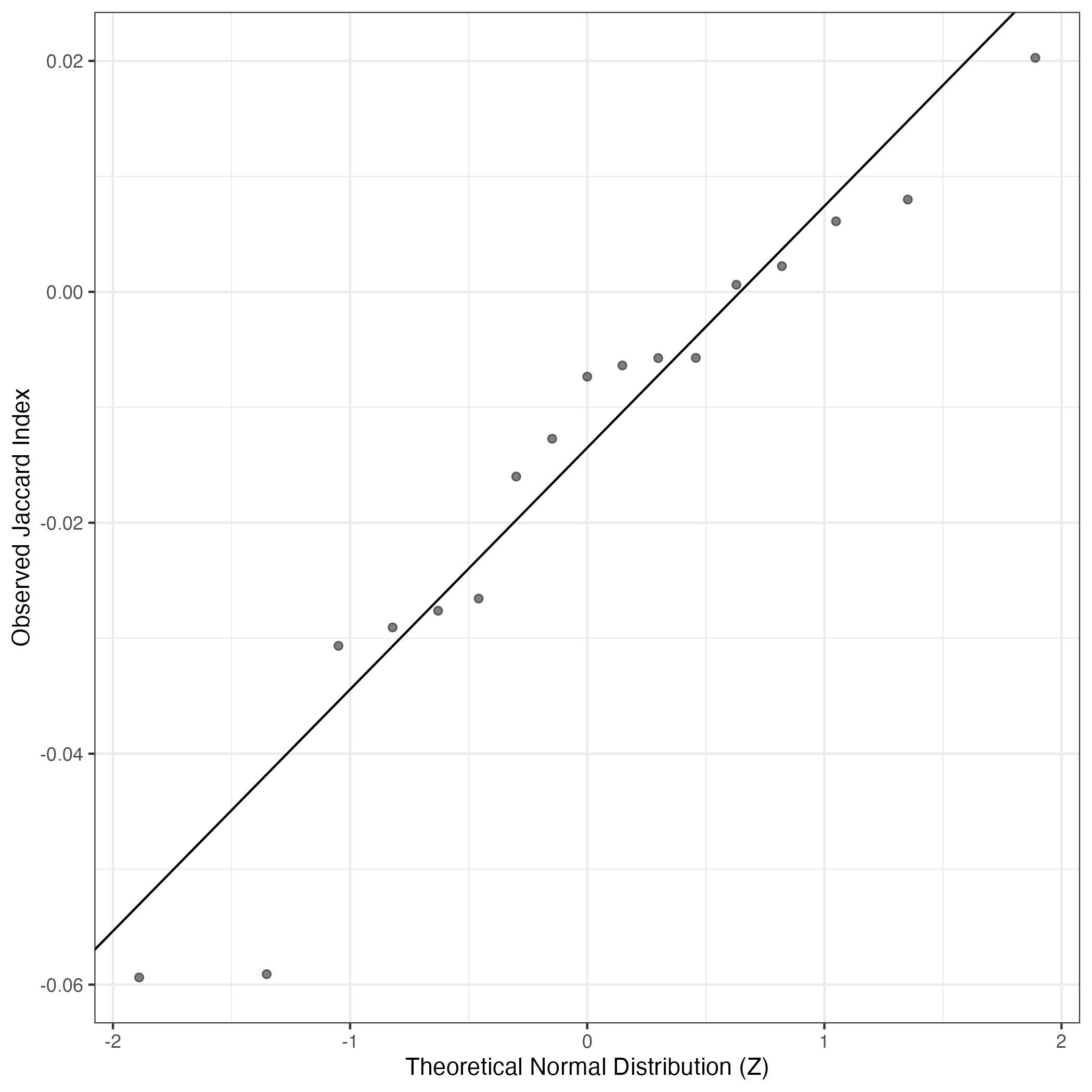
A similar QQ-plot as above, but only for instances where the H value is e.g. > 25, i.e. we are confident that the expression program is active above noise. Agreemenet between these binary vectors is tested using the Jaccard Index, with the P-values calculated by an exact test:
Interactive QQ-plot:
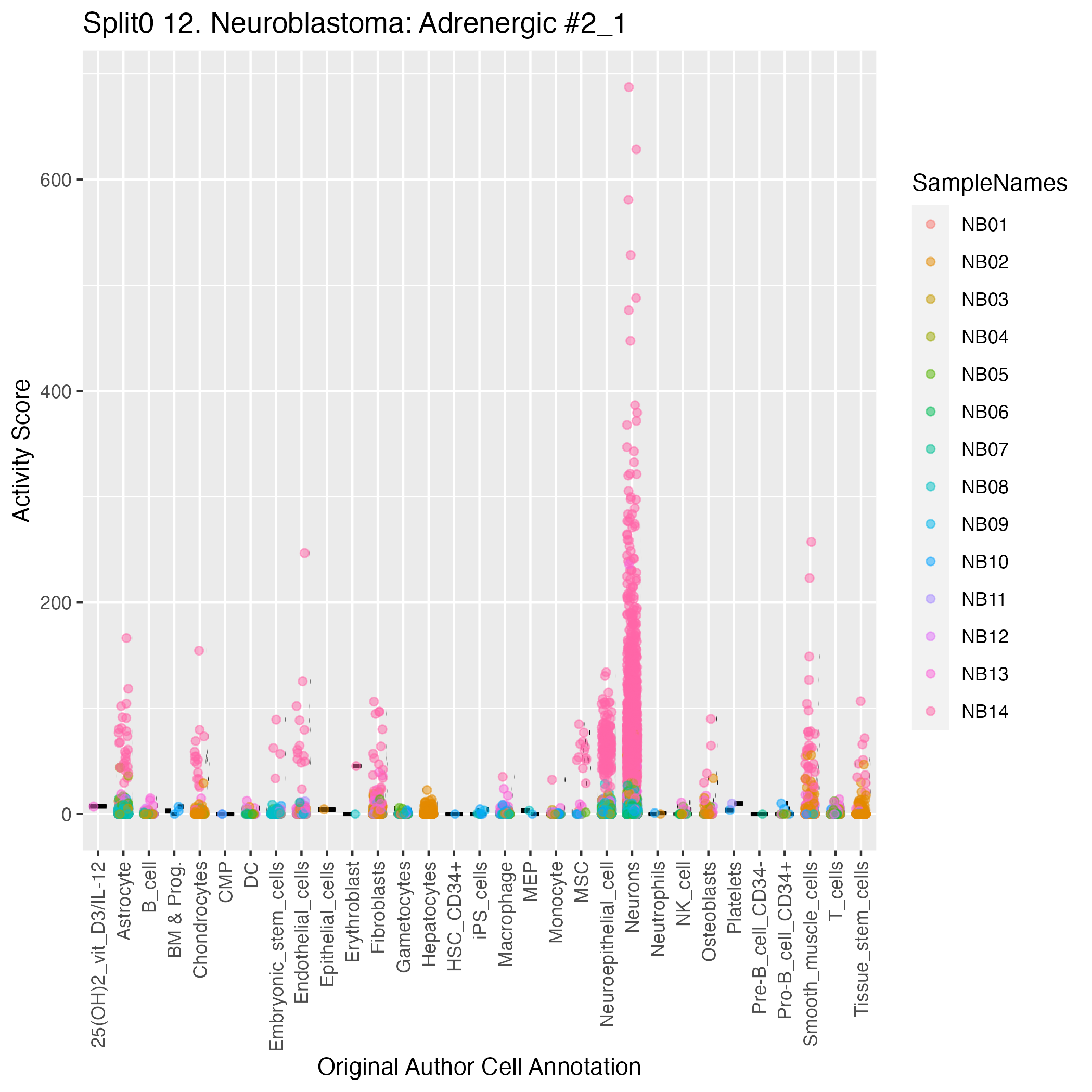
Singler cell type annotations for the top 50 cells on this program.
| Cell ID | Singler label | Singler Delta | Activity Score | Top Singler Raw Scores |
|---|---|---|---|---|
| NB14_TTGAACGGTTCTGAAC-1 | Neurons:adrenal_medulla_cell_line | 0.18 | 687.47 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.29, Neuroepithelial_cell:ESC-derived: 0.26, Astrocyte:Embryonic_stem_cell-derived: 0.25, Neurons:ES_cell-derived_neural_precursor: 0.25, MSC: 0.23, iPS_cells:CRL2097_foreskin: 0.22, Embryonic_stem_cells: 0.22, Smooth_muscle_cells:bronchial: 0.22, Tissue_stem_cells:CD326-CD56+: 0.22, iPS_cells:skin_fibroblast: 0.21 |
| NB14_CGCTATCGTACCGTAT-1 | Neurons:adrenal_medulla_cell_line | 0.20 | 628.66 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Neuroepithelial_cell:ESC-derived: 0.22, Astrocyte:Embryonic_stem_cell-derived: 0.22, Neurons:ES_cell-derived_neural_precursor: 0.21, iPS_cells:PDB_1lox-21Puro-20: 0.2, Tissue_stem_cells:CD326-CD56+: 0.2, Embryonic_stem_cells: 0.19, iPS_cells:PDB_1lox-21Puro-26: 0.19, iPS_cells:PDB_1lox-17Puro-5: 0.19, iPS_cells:PDB_1lox-17Puro-10: 0.19 |
| NB14_CAGATCATCCTACAGA-1 | Neurons:adrenal_medulla_cell_line | 0.18 | 580.86 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.26, Astrocyte:Embryonic_stem_cell-derived: 0.21, Neuroepithelial_cell:ESC-derived: 0.21, Neurons:ES_cell-derived_neural_precursor: 0.2, Tissue_stem_cells:CD326-CD56+: 0.18, Embryonic_stem_cells: 0.17, iPS_cells:PDB_1lox-21Puro-20: 0.17, iPS_cells:PDB_1lox-17Puro-10: 0.17, iPS_cells:PDB_1lox-21Puro-26: 0.16, iPS_cells:PDB_1lox-17Puro-5: 0.16 |
| NB14_ATAACGCTCTTTAGTC-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 528.58 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.28, Neuroepithelial_cell:ESC-derived: 0.24, Astrocyte:Embryonic_stem_cell-derived: 0.22, Neurons:ES_cell-derived_neural_precursor: 0.22, iPS_cells:PDB_1lox-21Puro-20: 0.22, iPS_cells:PDB_1lox-17Puro-10: 0.21, iPS_cells:PDB_1lox-21Puro-26: 0.21, Embryonic_stem_cells: 0.21, iPS_cells:PDB_1lox-17Puro-5: 0.21, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.21 |
| NB14_ATCATCTTCCAAATGC-1 | Neurons:adrenal_medulla_cell_line | 0.19 | 488.01 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.28, Neuroepithelial_cell:ESC-derived: 0.21, Astrocyte:Embryonic_stem_cell-derived: 0.21, Neurons:ES_cell-derived_neural_precursor: 0.19, iPS_cells:PDB_1lox-21Puro-20: 0.18, Embryonic_stem_cells: 0.18, iPS_cells:PDB_1lox-21Puro-26: 0.18, iPS_cells:PDB_1lox-17Puro-10: 0.17, iPS_cells:PDB_1lox-17Puro-5: 0.17, Tissue_stem_cells:CD326-CD56+: 0.17 |
| NB14_TTCTTAGAGTACGTAA-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 476.39 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.25, Neuroepithelial_cell:ESC-derived: 0.23, Astrocyte:Embryonic_stem_cell-derived: 0.22, Neurons:ES_cell-derived_neural_precursor: 0.21, Tissue_stem_cells:CD326-CD56+: 0.2, MSC: 0.2, Smooth_muscle_cells:bronchial: 0.2, Neurons:Schwann_cell: 0.2, Tissue_stem_cells:lipoma-derived_MSC: 0.2, Embryonic_stem_cells: 0.2 |
| NB14_AGGTCATAGTACCGGA-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 447.56 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.26, Neuroepithelial_cell:ESC-derived: 0.21, Astrocyte:Embryonic_stem_cell-derived: 0.2, Neurons:ES_cell-derived_neural_precursor: 0.2, iPS_cells:PDB_1lox-21Puro-20: 0.19, Tissue_stem_cells:CD326-CD56+: 0.19, iPS_cells:PDB_1lox-21Puro-26: 0.18, iPS_cells:PDB_1lox-17Puro-10: 0.18, iPS_cells:PDB_1lox-17Puro-5: 0.18, Embryonic_stem_cells: 0.18 |
| NB14_CCTACACCATACGCCG-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 386.65 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.25, Neuroepithelial_cell:ESC-derived: 0.2, Astrocyte:Embryonic_stem_cell-derived: 0.19, Neurons:ES_cell-derived_neural_precursor: 0.18, iPS_cells:PDB_1lox-21Puro-20: 0.18, iPS_cells:PDB_1lox-21Puro-26: 0.18, iPS_cells:PDB_1lox-17Puro-5: 0.18, iPS_cells:PDB_1lox-17Puro-10: 0.18, Embryonic_stem_cells: 0.17, iPS_cells:PDB_2lox-22: 0.17 |
| NB14_TACCTATGTCAACTGT-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 379.59 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.27, Neuroepithelial_cell:ESC-derived: 0.22, Neurons:ES_cell-derived_neural_precursor: 0.22, Astrocyte:Embryonic_stem_cell-derived: 0.21, Embryonic_stem_cells: 0.19, iPS_cells:PDB_1lox-21Puro-20: 0.19, iPS_cells:PDB_1lox-17Puro-10: 0.19, iPS_cells:PDB_1lox-21Puro-26: 0.19, iPS_cells:PDB_1lox-17Puro-5: 0.19, Tissue_stem_cells:CD326-CD56+: 0.19 |
| NB14_GTTCTCGCAAGAAAGG-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 371.83 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.22, Neuroepithelial_cell:ESC-derived: 0.17, Neurons:ES_cell-derived_neural_precursor: 0.16, Astrocyte:Embryonic_stem_cell-derived: 0.16, iPS_cells:PDB_1lox-21Puro-26: 0.16, iPS_cells:PDB_1lox-17Puro-10: 0.16, iPS_cells:PDB_1lox-21Puro-20: 0.16, iPS_cells:PDB_1lox-17Puro-5: 0.15, iPS_cells:PDB_2lox-22: 0.15, iPS_cells:PDB_2lox-21: 0.15 |
| NB14_CTCTAATCATCCTTGC-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 367.96 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.26, Neuroepithelial_cell:ESC-derived: 0.19, Astrocyte:Embryonic_stem_cell-derived: 0.18, Neurons:ES_cell-derived_neural_precursor: 0.18, iPS_cells:PDB_1lox-21Puro-20: 0.16, iPS_cells:PDB_1lox-21Puro-26: 0.16, Embryonic_stem_cells: 0.16, iPS_cells:PDB_1lox-17Puro-10: 0.16, iPS_cells:PDB_1lox-17Puro-5: 0.16, iPS_cells:PDB_2lox-22: 0.16 |
| NB14_AGCTCTCAGACGACGT-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 346.97 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.25, Neuroepithelial_cell:ESC-derived: 0.2, Astrocyte:Embryonic_stem_cell-derived: 0.2, Neurons:ES_cell-derived_neural_precursor: 0.19, Tissue_stem_cells:CD326-CD56+: 0.18, MSC: 0.17, Embryonic_stem_cells: 0.17, iPS_cells:PDB_1lox-21Puro-20: 0.17, Smooth_muscle_cells:vascular: 0.17, iPS_cells:PDB_1lox-21Puro-26: 0.17 |
| NB14_CTTGGCTGTAGCTAAA-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 343.08 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.26, Astrocyte:Embryonic_stem_cell-derived: 0.19, Neuroepithelial_cell:ESC-derived: 0.19, Neurons:ES_cell-derived_neural_precursor: 0.18, iPS_cells:PDB_1lox-17Puro-10: 0.17, iPS_cells:PDB_1lox-21Puro-20: 0.17, iPS_cells:PDB_1lox-17Puro-5: 0.17, Tissue_stem_cells:CD326-CD56+: 0.17, iPS_cells:PDB_1lox-21Puro-26: 0.16, Embryonic_stem_cells: 0.16 |
| NB14_TATGCCCAGATGCCTT-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 332.69 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.25, Neuroepithelial_cell:ESC-derived: 0.2, Astrocyte:Embryonic_stem_cell-derived: 0.19, Neurons:ES_cell-derived_neural_precursor: 0.18, iPS_cells:PDB_1lox-21Puro-20: 0.17, iPS_cells:PDB_1lox-21Puro-26: 0.17, iPS_cells:PDB_1lox-17Puro-5: 0.17, iPS_cells:PDB_1lox-17Puro-10: 0.17, Tissue_stem_cells:CD326-CD56+: 0.17, iPS_cells:PDB_2lox-22: 0.17 |
| NB14_ATTTCTGCATATGAGA-1 | Neurons:adrenal_medulla_cell_line | 0.18 | 321.81 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.25, Neuroepithelial_cell:ESC-derived: 0.19, Astrocyte:Embryonic_stem_cell-derived: 0.18, Neurons:ES_cell-derived_neural_precursor: 0.18, Tissue_stem_cells:CD326-CD56+: 0.16, Embryonic_stem_cells: 0.16, iPS_cells:PDB_1lox-21Puro-20: 0.16, iPS_cells:PDB_1lox-17Puro-10: 0.16, iPS_cells:PDB_1lox-21Puro-26: 0.16, iPS_cells:PDB_1lox-17Puro-5: 0.16 |
| NB14_TACCTATTCCGCTGTT-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 321.37 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.24, Neuroepithelial_cell:ESC-derived: 0.18, Astrocyte:Embryonic_stem_cell-derived: 0.18, Neurons:ES_cell-derived_neural_precursor: 0.17, Embryonic_stem_cells: 0.15, iPS_cells:PDB_1lox-17Puro-10: 0.15, iPS_cells:PDB_1lox-17Puro-5: 0.15, Tissue_stem_cells:CD326-CD56+: 0.15, iPS_cells:PDB_1lox-21Puro-26: 0.15, iPS_cells:PDB_1lox-21Puro-20: 0.15 |
| NB14_GACGGCTTCCTCCTAG-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 320.17 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.22, Neuroepithelial_cell:ESC-derived: 0.17, Neurons:ES_cell-derived_neural_precursor: 0.17, Astrocyte:Embryonic_stem_cell-derived: 0.17, Tissue_stem_cells:CD326-CD56+: 0.14, Embryonic_stem_cells: 0.14, iPS_cells:PDB_1lox-17Puro-5: 0.14, iPS_cells:PDB_1lox-21Puro-20: 0.14, iPS_cells:PDB_1lox-21Puro-26: 0.13, iPS_cells:PDB_1lox-17Puro-10: 0.13 |
| NB14_ATCCGAATCCGCGCAA-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 305.50 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.21, Neuroepithelial_cell:ESC-derived: 0.16, Astrocyte:Embryonic_stem_cell-derived: 0.15, Neurons:ES_cell-derived_neural_precursor: 0.15, Embryonic_stem_cells: 0.14, iPS_cells:PDB_1lox-21Puro-20: 0.14, Tissue_stem_cells:CD326-CD56+: 0.14, iPS_cells:PDB_1lox-21Puro-26: 0.14, iPS_cells:PDB_2lox-22: 0.14, iPS_cells:PDB_1lox-17Puro-5: 0.13 |
| NB14_AAACGGGAGGCGATAC-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 299.87 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.25, Neuroepithelial_cell:ESC-derived: 0.21, Neurons:ES_cell-derived_neural_precursor: 0.2, Astrocyte:Embryonic_stem_cell-derived: 0.19, Tissue_stem_cells:CD326-CD56+: 0.17, Neurons:Schwann_cell: 0.17, iPS_cells:PDB_1lox-17Puro-10: 0.17, iPS_cells:PDB_1lox-21Puro-20: 0.17, iPS_cells:PDB_1lox-17Puro-5: 0.17, Embryonic_stem_cells: 0.17 |
| NB14_TCTGGAATCTGCCCTA-1 | Fibroblasts:breast | 0.11 | 297.70 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.19, Neuroepithelial_cell:ESC-derived: 0.17, MSC: 0.17, Fibroblasts:breast: 0.16, Chondrocytes:MSC-derived: 0.16, Astrocyte:Embryonic_stem_cell-derived: 0.16, iPS_cells:CRL2097_foreskin: 0.16, iPS_cells:adipose_stem_cells: 0.16, Neurons:ES_cell-derived_neural_precursor: 0.16, Osteoblasts: 0.16 |
| NB14_CAGATCAGTGACCAAG-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 297.43 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.22, Astrocyte:Embryonic_stem_cell-derived: 0.17, Neuroepithelial_cell:ESC-derived: 0.16, Neurons:ES_cell-derived_neural_precursor: 0.16, iPS_cells:PDB_1lox-17Puro-10: 0.14, Neurons:Schwann_cell: 0.14, Embryonic_stem_cells: 0.14, Tissue_stem_cells:CD326-CD56+: 0.14, iPS_cells:PDB_1lox-21Puro-26: 0.14, iPS_cells:PDB_1lox-17Puro-5: 0.14 |
| NB14_TTTGGTTGTAAATGAC-1 | Neurons:adrenal_medulla_cell_line | 0.12 | 289.36 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.2, Neuroepithelial_cell:ESC-derived: 0.16, Astrocyte:Embryonic_stem_cell-derived: 0.15, Neurons:ES_cell-derived_neural_precursor: 0.15, iPS_cells:PDB_1lox-17Puro-5: 0.14, iPS_cells:PDB_1lox-21Puro-20: 0.14, iPS_cells:PDB_1lox-17Puro-10: 0.14, iPS_cells:PDB_1lox-21Puro-26: 0.14, Embryonic_stem_cells: 0.14, iPS_cells:PDB_2lox-5: 0.14 |
| NB14_AGTGGGATCTGAGGGA-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 283.69 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.22, Neuroepithelial_cell:ESC-derived: 0.18, Neurons:ES_cell-derived_neural_precursor: 0.17, Astrocyte:Embryonic_stem_cell-derived: 0.17, iPS_cells:PDB_1lox-21Puro-26: 0.16, iPS_cells:PDB_1lox-21Puro-20: 0.16, iPS_cells:PDB_1lox-17Puro-5: 0.16, iPS_cells:PDB_1lox-17Puro-10: 0.16, Embryonic_stem_cells: 0.15, iPS_cells:PDB_2lox-5: 0.15 |
| NB14_CAGATCAGTTTAGCTG-1 | Neurons:adrenal_medulla_cell_line | 0.11 | 283.55 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.2, Neuroepithelial_cell:ESC-derived: 0.15, Astrocyte:Embryonic_stem_cell-derived: 0.15, Tissue_stem_cells:CD326-CD56+: 0.14, Endothelial_cells:HUVEC:VEGF: 0.14, Neurons:ES_cell-derived_neural_precursor: 0.14, Chondrocytes:MSC-derived: 0.14, Endothelial_cells:HUVEC: 0.13, Endothelial_cells:lymphatic: 0.13, Neurons:Schwann_cell: 0.13 |
| NB14_GACTACAGTAGTAGTA-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 279.79 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.25, Neuroepithelial_cell:ESC-derived: 0.21, Astrocyte:Embryonic_stem_cell-derived: 0.21, Neurons:ES_cell-derived_neural_precursor: 0.2, Embryonic_stem_cells: 0.19, iPS_cells:PDB_1lox-21Puro-20: 0.19, iPS_cells:PDB_1lox-21Puro-26: 0.19, iPS_cells:PDB_1lox-17Puro-10: 0.19, iPS_cells:PDB_1lox-17Puro-5: 0.19, iPS_cells:PDB_2lox-22: 0.19 |
| NB14_ACTTGTTCAGCTGCTG-1 | Neurons:adrenal_medulla_cell_line | 0.11 | 277.13 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.16, Neurons:ES_cell-derived_neural_precursor: 0.14, Neuroepithelial_cell:ESC-derived: 0.14, iPS_cells:PDB_1lox-17Puro-5: 0.13, iPS_cells:PDB_1lox-17Puro-10: 0.13, iPS_cells:PDB_1lox-21Puro-20: 0.13, iPS_cells:PDB_1lox-21Puro-26: 0.13, Embryonic_stem_cells: 0.13, Astrocyte:Embryonic_stem_cell-derived: 0.13, iPS_cells:PDB_2lox-22: 0.12 |
| NB14_AGGGATGCAGACAAAT-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 276.59 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.24, Neuroepithelial_cell:ESC-derived: 0.18, Neurons:ES_cell-derived_neural_precursor: 0.18, Astrocyte:Embryonic_stem_cell-derived: 0.17, Embryonic_stem_cells: 0.16, iPS_cells:PDB_1lox-21Puro-20: 0.16, iPS_cells:PDB_2lox-22: 0.15, iPS_cells:PDB_1lox-17Puro-10: 0.15, iPS_cells:PDB_1lox-17Puro-5: 0.15, iPS_cells:PDB_1lox-21Puro-26: 0.15 |
| NB14_GGACAAGCATCTCGCT-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 274.50 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.21, Neuroepithelial_cell:ESC-derived: 0.17, Neurons:ES_cell-derived_neural_precursor: 0.16, Astrocyte:Embryonic_stem_cell-derived: 0.16, iPS_cells:PDB_1lox-21Puro-20: 0.15, Tissue_stem_cells:CD326-CD56+: 0.15, iPS_cells:PDB_1lox-17Puro-5: 0.15, iPS_cells:PDB_1lox-21Puro-26: 0.15, iPS_cells:PDB_1lox-17Puro-10: 0.15, Embryonic_stem_cells: 0.14 |
| NB14_CGGACTGGTGCACCAC-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 272.14 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.2, Neuroepithelial_cell:ESC-derived: 0.14, Astrocyte:Embryonic_stem_cell-derived: 0.14, Neurons:ES_cell-derived_neural_precursor: 0.13, iPS_cells:PDB_1lox-21Puro-20: 0.13, iPS_cells:PDB_1lox-17Puro-5: 0.13, iPS_cells:PDB_1lox-17Puro-10: 0.13, Embryonic_stem_cells: 0.13, iPS_cells:PDB_1lox-21Puro-26: 0.13, Tissue_stem_cells:CD326-CD56+: 0.12 |
| NB14_CTGGTCTAGCACACAG-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 271.02 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.22, Neuroepithelial_cell:ESC-derived: 0.18, Neurons:ES_cell-derived_neural_precursor: 0.18, Astrocyte:Embryonic_stem_cell-derived: 0.17, iPS_cells:PDB_1lox-17Puro-10: 0.16, iPS_cells:PDB_1lox-17Puro-5: 0.16, Tissue_stem_cells:CD326-CD56+: 0.16, iPS_cells:PDB_1lox-21Puro-26: 0.16, iPS_cells:PDB_1lox-21Puro-20: 0.16, iPS_cells:PDB_2lox-22: 0.16 |
| NB14_ATCACGATCATGTGGT-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 264.57 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.22, Neuroepithelial_cell:ESC-derived: 0.16, Astrocyte:Embryonic_stem_cell-derived: 0.16, Neurons:ES_cell-derived_neural_precursor: 0.16, Tissue_stem_cells:CD326-CD56+: 0.15, iPS_cells:PDB_1lox-21Puro-20: 0.14, Embryonic_stem_cells: 0.14, iPS_cells:PDB_1lox-21Puro-26: 0.14, iPS_cells:PDB_1lox-17Puro-5: 0.14, iPS_cells:skin_fibroblast-derived: 0.14 |
| NB14_GTGCATAAGTTCCACA-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 263.78 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.22, Neuroepithelial_cell:ESC-derived: 0.17, Astrocyte:Embryonic_stem_cell-derived: 0.15, Neurons:ES_cell-derived_neural_precursor: 0.15, Tissue_stem_cells:CD326-CD56+: 0.14, iPS_cells:PDB_1lox-17Puro-5: 0.14, iPS_cells:PDB_1lox-17Puro-10: 0.14, iPS_cells:PDB_1lox-21Puro-26: 0.14, iPS_cells:PDB_1lox-21Puro-20: 0.14, Embryonic_stem_cells: 0.14 |
| NB14_AAAGATGTCGGCATCG-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 259.20 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.22, Neuroepithelial_cell:ESC-derived: 0.19, Astrocyte:Embryonic_stem_cell-derived: 0.17, Neurons:ES_cell-derived_neural_precursor: 0.17, Tissue_stem_cells:CD326-CD56+: 0.16, Embryonic_stem_cells: 0.16, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.15, MSC: 0.15, iPS_cells:PDB_1lox-21Puro-26: 0.15, iPS_cells:PDB_1lox-21Puro-20: 0.15 |
| NB14_CTAGCCTTCACTATTC-1 | Neurons:adrenal_medulla_cell_line | 0.13 | 258.93 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.2, Neuroepithelial_cell:ESC-derived: 0.16, Astrocyte:Embryonic_stem_cell-derived: 0.16, Neurons:ES_cell-derived_neural_precursor: 0.15, iPS_cells:PDB_1lox-21Puro-20: 0.14, iPS_cells:PDB_1lox-17Puro-10: 0.14, iPS_cells:PDB_1lox-21Puro-26: 0.14, iPS_cells:PDB_1lox-17Puro-5: 0.14, Embryonic_stem_cells: 0.14, iPS_cells:skin_fibroblast-derived: 0.14 |
| NB14_TCCACACAGTTTAGGA-1 | Neurons:adrenal_medulla_cell_line | 0.13 | 257.38 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.2, Neuroepithelial_cell:ESC-derived: 0.18, Smooth_muscle_cells:bronchial: 0.18, Smooth_muscle_cells:bronchial:vit_D: 0.17, MSC: 0.17, Neurons:ES_cell-derived_neural_precursor: 0.17, iPS_cells:CRL2097_foreskin: 0.17, Tissue_stem_cells:CD326-CD56+: 0.17, iPS_cells:skin_fibroblast: 0.17, Fibroblasts:breast: 0.17 |
| NB14_CACAAACTCTCCGGTT-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 253.26 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.21, Neuroepithelial_cell:ESC-derived: 0.16, Neurons:ES_cell-derived_neural_precursor: 0.16, Astrocyte:Embryonic_stem_cell-derived: 0.15, Embryonic_stem_cells: 0.14, Tissue_stem_cells:CD326-CD56+: 0.13, iPS_cells:PDB_1lox-21Puro-20: 0.13, iPS_cells:PDB_1lox-17Puro-10: 0.13, iPS_cells:PDB_1lox-17Puro-5: 0.13, iPS_cells:PDB_1lox-21Puro-26: 0.13 |
| NB14_CGTCAGGTCCAAACAC-1 | Neurons:adrenal_medulla_cell_line | 0.12 | 248.63 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.19, Astrocyte:Embryonic_stem_cell-derived: 0.12, Neuroepithelial_cell:ESC-derived: 0.12, iPS_cells:PDB_1lox-21Puro-26: 0.12, iPS_cells:PDB_1lox-21Puro-20: 0.11, iPS_cells:PDB_1lox-17Puro-5: 0.11, iPS_cells:PDB_1lox-17Puro-10: 0.11, Embryonic_stem_cells: 0.11, Tissue_stem_cells:CD326-CD56+: 0.11, Neurons:ES_cell-derived_neural_precursor: 0.11 |
| NB14_ATGAGGGTCCCGGATG-1 | Endothelial_cells:lymphatic:KSHV | 0.16 | 246.77 | Raw ScoresEndothelial_cells:lymphatic: 0.3, Endothelial_cells:HUVEC:VEGF: 0.3, Endothelial_cells:HUVEC:Serum_Amyloid_A: 0.29, Endothelial_cells:lymphatic:KSHV: 0.29, Endothelial_cells:lymphatic:TNFa_48h: 0.29, Endothelial_cells:HUVEC: 0.29, Endothelial_cells:blood_vessel: 0.29, Endothelial_cells:HUVEC:IL-1b: 0.27, Endothelial_cells:HUVEC:B._anthracis_LT: 0.27, iPS_cells:CRL2097_foreskin: 0.27 |
| NB14_GTAACGTCACCAGATT-1 | Neurons:adrenal_medulla_cell_line | 0.11 | 244.46 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.27, Neuroepithelial_cell:ESC-derived: 0.23, Astrocyte:Embryonic_stem_cell-derived: 0.22, Neurons:ES_cell-derived_neural_precursor: 0.22, iPS_cells:PDB_1lox-21Puro-26: 0.21, iPS_cells:PDB_2lox-22: 0.21, iPS_cells:PDB_1lox-17Puro-10: 0.21, iPS_cells:PDB_1lox-21Puro-20: 0.21, iPS_cells:PDB_1lox-17Puro-5: 0.21, iPS_cells:PDB_2lox-5: 0.21 |
| NB14_TTGACTTCAATCTACG-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 242.03 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.26, Neuroepithelial_cell:ESC-derived: 0.2, Neurons:ES_cell-derived_neural_precursor: 0.19, Tissue_stem_cells:CD326-CD56+: 0.18, Astrocyte:Embryonic_stem_cell-derived: 0.18, MSC: 0.17, Endothelial_cells:HUVEC:VEGF: 0.17, Neurons:Schwann_cell: 0.17, iPS_cells:PDB_1lox-17Puro-5: 0.17, iPS_cells:PDB_1lox-21Puro-20: 0.17 |
| NB14_CTGATCCAGGATGGAA-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 241.35 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.21, Neuroepithelial_cell:ESC-derived: 0.16, Astrocyte:Embryonic_stem_cell-derived: 0.14, Neurons:ES_cell-derived_neural_precursor: 0.14, MSC: 0.13, Neurons:Schwann_cell: 0.13, iPS_cells:PDB_1lox-17Puro-10: 0.13, Tissue_stem_cells:CD326-CD56+: 0.13, iPS_cells:PDB_1lox-21Puro-20: 0.13, iPS_cells:PDB_1lox-21Puro-26: 0.13 |
| NB14_CTAGTGAAGCTGAACG-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 239.63 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.22, Neuroepithelial_cell:ESC-derived: 0.18, Neurons:ES_cell-derived_neural_precursor: 0.18, Astrocyte:Embryonic_stem_cell-derived: 0.17, Embryonic_stem_cells: 0.16, iPS_cells:PDB_1lox-21Puro-20: 0.16, iPS_cells:PDB_1lox-17Puro-10: 0.16, iPS_cells:PDB_1lox-21Puro-26: 0.16, iPS_cells:PDB_1lox-17Puro-5: 0.16, iPS_cells:skin_fibroblast-derived: 0.15 |
| NB14_GGATTACGTAGCGATG-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 238.88 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.25, Neuroepithelial_cell:ESC-derived: 0.2, Astrocyte:Embryonic_stem_cell-derived: 0.19, Neurons:ES_cell-derived_neural_precursor: 0.19, Embryonic_stem_cells: 0.18, Tissue_stem_cells:CD326-CD56+: 0.18, iPS_cells:PDB_1lox-21Puro-20: 0.17, iPS_cells:PDB_1lox-17Puro-10: 0.17, iPS_cells:PDB_2lox-22: 0.17, iPS_cells:PDB_1lox-21Puro-26: 0.17 |
| NB13_CTAACTTAGCCGCCTA-1 | Neurons:adrenal_medulla_cell_line | 0.23 | 236.23 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.42, Astrocyte:Embryonic_stem_cell-derived: 0.34, Neuroepithelial_cell:ESC-derived: 0.33, Neurons:ES_cell-derived_neural_precursor: 0.31, iPS_cells:PDB_1lox-21Puro-20: 0.3, iPS_cells:PDB_1lox-21Puro-26: 0.3, iPS_cells:PDB_1lox-17Puro-10: 0.3, iPS_cells:PDB_1lox-17Puro-5: 0.3, Embryonic_stem_cells: 0.29, iPS_cells:PDB_2lox-22: 0.29 |
| NB14_AAGGAGCAGAAACCGC-1 | Neurons:adrenal_medulla_cell_line | 0.10 | 230.74 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.18, Neuroepithelial_cell:ESC-derived: 0.15, Astrocyte:Embryonic_stem_cell-derived: 0.15, Neurons:ES_cell-derived_neural_precursor: 0.15, Chondrocytes:MSC-derived: 0.15, Smooth_muscle_cells:bronchial: 0.15, Osteoblasts: 0.15, Smooth_muscle_cells:bronchial:vit_D: 0.15, iPS_cells:CRL2097_foreskin: 0.14, Tissue_stem_cells:BM_MSC:BMP2: 0.14 |
| NB14_AGATTGCAGTCATCCA-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 230.32 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.21, Neuroepithelial_cell:ESC-derived: 0.16, Neurons:ES_cell-derived_neural_precursor: 0.15, Astrocyte:Embryonic_stem_cell-derived: 0.15, iPS_cells:PDB_1lox-21Puro-26: 0.14, iPS_cells:PDB_1lox-21Puro-20: 0.14, Tissue_stem_cells:CD326-CD56+: 0.14, iPS_cells:PDB_1lox-17Puro-5: 0.14, iPS_cells:PDB_1lox-17Puro-10: 0.14, Embryonic_stem_cells: 0.14 |
| NB14_TAAGCGTAGAGATGAG-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 230.31 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.26, Neuroepithelial_cell:ESC-derived: 0.19, Astrocyte:Embryonic_stem_cell-derived: 0.18, iPS_cells:PDB_1lox-21Puro-20: 0.17, Neurons:ES_cell-derived_neural_precursor: 0.17, iPS_cells:PDB_1lox-21Puro-26: 0.17, iPS_cells:PDB_1lox-17Puro-10: 0.17, Tissue_stem_cells:CD326-CD56+: 0.17, iPS_cells:PDB_1lox-17Puro-5: 0.17, iPS_cells:PDB_2lox-22: 0.17 |
| NB14_AAAGCAAAGGACAGAA-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 228.35 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.23, Neuroepithelial_cell:ESC-derived: 0.17, Neurons:ES_cell-derived_neural_precursor: 0.17, Astrocyte:Embryonic_stem_cell-derived: 0.17, Tissue_stem_cells:CD326-CD56+: 0.16, iPS_cells:PDB_1lox-17Puro-5: 0.16, iPS_cells:PDB_1lox-17Puro-10: 0.16, iPS_cells:PDB_1lox-21Puro-26: 0.16, iPS_cells:PDB_1lox-21Puro-20: 0.16, Embryonic_stem_cells: 0.15 |
| NB14_CGTGAGCGTTGAACTC-1 | Neurons:adrenal_medulla_cell_line | 0.13 | 224.79 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.21, Astrocyte:Embryonic_stem_cell-derived: 0.17, Neuroepithelial_cell:ESC-derived: 0.16, Neurons:ES_cell-derived_neural_precursor: 0.16, iPS_cells:PDB_1lox-21Puro-20: 0.15, iPS_cells:PDB_1lox-21Puro-26: 0.14, iPS_cells:PDB_1lox-17Puro-5: 0.14, iPS_cells:PDB_1lox-17Puro-10: 0.14, Embryonic_stem_cells: 0.14, Tissue_stem_cells:CD326-CD56+: 0.14 |
| NB14_GTAACTGAGTACGATA-1 | Neurons:adrenal_medulla_cell_line | 0.12 | 223.09 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.19, Neuroepithelial_cell:ESC-derived: 0.15, Smooth_muscle_cells:bronchial:vit_D: 0.15, Smooth_muscle_cells:bronchial: 0.15, Astrocyte:Embryonic_stem_cell-derived: 0.15, Smooth_muscle_cells:vascular:IL-17: 0.15, Smooth_muscle_cells:vascular: 0.14, Neurons:ES_cell-derived_neural_precursor: 0.14, iPS_cells:CRL2097_foreskin: 0.14, iPS_cells:adipose_stem_cells: 0.14 |
Below shows the significant enrichments of this GEP for literature curated gene lists
This data was procured from existing single cell RNA-seq maps of neuroblastoma or related relevant data.
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by the P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Myofibroblastic CAF
These marker genes were curated across cancer subtypes in multiple organ systems as reviewed in Lavie et. al. (PMID 35883004) and contain myofibroblastic specific CAF genes:
Wilcoxon ranksum test P-value for gene set overrepresentation: 7.85e-06
Mean rank of genes in gene set: 4044.37
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| RGS5 | 0.0044652 | 126 | GTEx | DepMap | Descartes | 0.58 | 439.02 |
| COL1A1 | 0.0036897 | 182 | GTEx | DepMap | Descartes | 0.71 | 457.45 |
| POSTN | 0.0036784 | 184 | GTEx | DepMap | Descartes | 0.18 | 227.92 |
| MMP2 | 0.0028062 | 329 | GTEx | DepMap | Descartes | 0.12 | 143.11 |
| COL1A2 | 0.0024567 | 412 | GTEx | DepMap | Descartes | 0.77 | 491.83 |
| ACTG2 | 0.0022909 | 462 | GTEx | DepMap | Descartes | 0.02 | 48.94 |
| PGF | 0.0021582 | 509 | GTEx | DepMap | Descartes | 0.06 | 61.96 |
| TPM2 | 0.0020322 | 569 | GTEx | DepMap | Descartes | 0.10 | 227.36 |
| FN1 | 0.0016008 | 851 | GTEx | DepMap | Descartes | 0.33 | 144.54 |
| COL3A1 | 0.0015482 | 893 | GTEx | DepMap | Descartes | 0.36 | 255.97 |
| IGFBP7 | 0.0012343 | 1239 | GTEx | DepMap | Descartes | 0.19 | 486.05 |
| BGN | 0.0012075 | 1289 | GTEx | DepMap | Descartes | 0.08 | 124.94 |
| CNN2 | 0.0010529 | 1512 | GTEx | DepMap | Descartes | 0.02 | 30.58 |
| COL5A1 | 0.0010088 | 1583 | GTEx | DepMap | Descartes | 0.09 | 38.63 |
| COL4A1 | 0.0009925 | 1613 | GTEx | DepMap | Descartes | 0.09 | 53.96 |
| COL15A1 | 0.0008872 | 1818 | GTEx | DepMap | Descartes | 0.03 | 17.18 |
| TGFB1 | 0.0008181 | 2013 | GTEx | DepMap | Descartes | 0.07 | 102.79 |
| THY1 | 0.0008003 | 2052 | GTEx | DepMap | Descartes | 0.06 | 57.35 |
| ACTA2 | 0.0007822 | 2098 | GTEx | DepMap | Descartes | 0.05 | 142.89 |
| WNT5A | 0.0007343 | 2242 | GTEx | DepMap | Descartes | 0.01 | 3.97 |
| TNC | 0.0006739 | 2440 | GTEx | DepMap | Descartes | 0.06 | 28.10 |
| COL5A2 | 0.0005867 | 2742 | GTEx | DepMap | Descartes | 0.10 | 53.22 |
| COL11A1 | 0.0005232 | 2990 | GTEx | DepMap | Descartes | 0.04 | 16.84 |
| MMP11 | 0.0005146 | 3030 | GTEx | DepMap | Descartes | 0.01 | 12.50 |
| TPM1 | 0.0004832 | 3172 | GTEx | DepMap | Descartes | 0.13 | 126.57 |
| DCN | 0.0004397 | 3385 | GTEx | DepMap | Descartes | 0.04 | 19.63 |
| COL13A1 | 0.0003946 | 3599 | GTEx | DepMap | Descartes | 0.01 | 11.68 |
| COL10A1 | 0.0003766 | 3708 | GTEx | DepMap | Descartes | 0.01 | 4.63 |
| MYL9 | 0.0003369 | 3948 | GTEx | DepMap | Descartes | 0.03 | 48.06 |
| VEGFA | 0.0003045 | 4134 | GTEx | DepMap | Descartes | 0.06 | 19.32 |
| LUM | 0.0002989 | 4176 | GTEx | DepMap | Descartes | 0.05 | 64.58 |
| CNN3 | 0.0002737 | 4325 | GTEx | DepMap | Descartes | 0.07 | 151.37 |
| THBS2 | 0.0002528 | 4470 | GTEx | DepMap | Descartes | 0.05 | 32.34 |
| TAGLN | 0.0002194 | 4692 | GTEx | DepMap | Descartes | 0.03 | 37.32 |
| TMEM119 | 0.0001538 | 5174 | GTEx | DepMap | Descartes | 0.01 | 11.07 |
| TGFBR1 | 0.0000703 | 5828 | GTEx | DepMap | Descartes | 0.08 | 49.59 |
| COL12A1 | 0.0000571 | 5935 | GTEx | DepMap | Descartes | 0.03 | 10.04 |
| TGFB2 | -0.0000508 | 6950 | GTEx | DepMap | Descartes | 0.01 | 5.05 |
| COL14A1 | -0.0000725 | 7230 | GTEx | DepMap | Descartes | 0.02 | 8.10 |
| IGFBP3 | -0.0000761 | 7274 | GTEx | DepMap | Descartes | 0.01 | 16.09 |
| HOPX | -0.0001071 | 7641 | GTEx | DepMap | Descartes | 0.00 | 2.59 |
| MYLK | -0.0001620 | 8257 | GTEx | DepMap | Descartes | 0.01 | 2.75 |
| ITGA7 | -0.0002172 | 8795 | GTEx | DepMap | Descartes | 0.01 | 4.07 |
| COL8A1 | -0.0002342 | 8959 | GTEx | DepMap | Descartes | 0.03 | 17.44 |
| MYH11 | -0.0002928 | 9432 | GTEx | DepMap | Descartes | 0.00 | 1.74 |
| TGFBR2 | -0.0004236 | 10313 | GTEx | DepMap | Descartes | 0.02 | 9.92 |
| MEF2C | -0.0004400 | 10416 | GTEx | DepMap | Descartes | 0.04 | 16.58 |
| VCAN | -0.0007282 | 11456 | GTEx | DepMap | Descartes | 0.06 | 16.35 |
| THBS1 | -0.0008695 | 11727 | GTEx | DepMap | Descartes | 0.03 | 13.44 |
Mesenchymal Fig 1D (Olsen)
Selected mesenchymal marker genes shown in Fig. 1D of Olsen et al. https://www.biorxiv.org/content/10.1101/2020.05.04.077057v1 - these are highly expressed in their mesenchymal cluster on their UMAP.:
Wilcoxon ranksum test P-value for gene set overrepresentation: 8.10e-04
Mean rank of genes in gene set: 3109.23
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| COL1A1 | 0.0036897 | 182 | GTEx | DepMap | Descartes | 0.71 | 457.45 |
| COL1A2 | 0.0024567 | 412 | GTEx | DepMap | Descartes | 0.77 | 491.83 |
| SPARC | 0.0021406 | 517 | GTEx | DepMap | Descartes | 0.27 | 314.80 |
| CALD1 | 0.0021119 | 533 | GTEx | DepMap | Descartes | 0.46 | 344.84 |
| COL3A1 | 0.0015482 | 893 | GTEx | DepMap | Descartes | 0.36 | 255.97 |
| COL6A2 | 0.0014929 | 943 | GTEx | DepMap | Descartes | 0.16 | 152.72 |
| BGN | 0.0012075 | 1289 | GTEx | DepMap | Descartes | 0.08 | 124.94 |
| PRRX1 | 0.0007719 | 2122 | GTEx | DepMap | Descartes | 0.03 | 27.69 |
| DCN | 0.0004397 | 3385 | GTEx | DepMap | Descartes | 0.04 | 19.63 |
| LUM | 0.0002989 | 4176 | GTEx | DepMap | Descartes | 0.05 | 64.58 |
| PDGFRA | -0.0000122 | 6540 | GTEx | DepMap | Descartes | 0.01 | 3.30 |
| LEPR | -0.0001676 | 8328 | GTEx | DepMap | Descartes | 0.02 | 9.09 |
| MGP | -0.0006001 | 11100 | GTEx | DepMap | Descartes | 0.03 | 67.34 |
Mesenchymal cells (Jansky)
Mentioned in the main text (Jansky et al, Nature Genetics (2021)), page 1, that COL1A1+ Mesenchymal were identified in their fetal adrenal glands, this is also highlighted in the UMAP plot on their Fig 1B. Additional genes also shown in their Extended data Figure 2D.:
Wilcoxon ranksum test P-value for gene set overrepresentation: 4.47e-03
Mean rank of genes in gene set: 808
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| COL1A1 | 0.0036897 | 182 | GTEx | DepMap | Descartes | 0.71 | 457.45 |
| COL1A2 | 0.0024567 | 412 | GTEx | DepMap | Descartes | 0.77 | 491.83 |
| PDGFRB | 0.0008819 | 1830 | GTEx | DepMap | Descartes | 0.03 | 18.87 |
Below shows ranks on this GEP for literature curated gene lists for large gene sets
These include those reported as mesenchymal/adrenergic by Van Groningen et al.
High ranks indicate this gene is a driver of this GEP (note these results are not ordered).
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
VanGroningen Adrenergic Genes
Adrenergic marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 7006.35
Median rank of genes in gene set: 7774
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CHGA | 0.0197936 | 1 | GTEx | DepMap | Descartes | 3.32 | 6803.85 |
| DLK1 | 0.0169486 | 3 | GTEx | DepMap | Descartes | 1.54 | 1490.73 |
| STRA6 | 0.0169388 | 5 | GTEx | DepMap | Descartes | 0.80 | 1103.42 |
| FAM155A | 0.0150706 | 7 | GTEx | DepMap | Descartes | 5.30 | 2348.91 |
| RET | 0.0145396 | 8 | GTEx | DepMap | Descartes | 0.58 | 405.80 |
| RRM2 | 0.0116163 | 16 | GTEx | DepMap | Descartes | 0.60 | 718.97 |
| CACNA2D2 | 0.0090320 | 26 | GTEx | DepMap | Descartes | 0.28 | 198.57 |
| CHGB | 0.0073304 | 46 | GTEx | DepMap | Descartes | 1.18 | 2045.75 |
| PRSS12 | 0.0061842 | 60 | GTEx | DepMap | Descartes | 0.24 | 199.08 |
| MSI2 | 0.0049908 | 91 | GTEx | DepMap | Descartes | 1.07 | 523.05 |
| THSD7A | 0.0049290 | 97 | GTEx | DepMap | Descartes | 1.23 | 478.14 |
| MAGI3 | 0.0045626 | 120 | GTEx | DepMap | Descartes | 0.49 | 283.49 |
| RGS5 | 0.0044652 | 126 | GTEx | DepMap | Descartes | 0.58 | 439.02 |
| DACH1 | 0.0042393 | 141 | GTEx | DepMap | Descartes | 0.43 | 363.75 |
| GATA2 | 0.0041690 | 146 | GTEx | DepMap | Descartes | 0.35 | 419.79 |
| ESRRG | 0.0041168 | 151 | GTEx | DepMap | Descartes | 0.32 | 237.09 |
| HAND1 | 0.0037780 | 174 | GTEx | DepMap | Descartes | 0.17 | 428.98 |
| CACNA1B | 0.0037575 | 175 | GTEx | DepMap | Descartes | 0.67 | 241.66 |
| PHF21B | 0.0037106 | 180 | GTEx | DepMap | Descartes | 0.18 | 164.18 |
| RBMS3 | 0.0035984 | 195 | GTEx | DepMap | Descartes | 1.93 | 889.71 |
| HEY1 | 0.0035780 | 200 | GTEx | DepMap | Descartes | 0.05 | 60.81 |
| CEP44 | 0.0034873 | 213 | GTEx | DepMap | Descartes | 0.20 | 207.51 |
| DPYSL3 | 0.0034713 | 216 | GTEx | DepMap | Descartes | 0.45 | 343.52 |
| EML4 | 0.0034606 | 217 | GTEx | DepMap | Descartes | 0.37 | 272.18 |
| RALGDS | 0.0032220 | 250 | GTEx | DepMap | Descartes | 0.28 | 171.82 |
| EML6 | 0.0031882 | 257 | GTEx | DepMap | Descartes | 0.42 | 169.87 |
| CYGB | 0.0031540 | 262 | GTEx | DepMap | Descartes | 0.16 | 315.92 |
| INSM2 | 0.0031321 | 264 | GTEx | DepMap | Descartes | 0.17 | 241.48 |
| SHC3 | 0.0029418 | 297 | GTEx | DepMap | Descartes | 0.12 | 47.60 |
| IRS2 | 0.0029187 | 303 | GTEx | DepMap | Descartes | 0.19 | 93.71 |
VanGroningen Mesenchymal Genes
Mesenchymal marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.72e-06
Mean rank of genes in gene set: 5482.7
Median rank of genes in gene set: 4824
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| PDIA6 | 0.0169478 | 4 | GTEx | DepMap | Descartes | 1.76 | 2883.91 |
| ITPR1 | 0.0069989 | 51 | GTEx | DepMap | Descartes | 0.22 | 89.16 |
| SHC1 | 0.0054258 | 79 | GTEx | DepMap | Descartes | 0.36 | 415.95 |
| CRABP2 | 0.0051622 | 85 | GTEx | DepMap | Descartes | 0.09 | 376.97 |
| SPRY4 | 0.0048408 | 101 | GTEx | DepMap | Descartes | 0.07 | 57.15 |
| ACADVL | 0.0047749 | 107 | GTEx | DepMap | Descartes | 0.32 | 496.53 |
| SHROOM3 | 0.0043774 | 133 | GTEx | DepMap | Descartes | 0.09 | 33.58 |
| FLNA | 0.0043616 | 134 | GTEx | DepMap | Descartes | 0.26 | 120.95 |
| COL1A1 | 0.0036897 | 182 | GTEx | DepMap | Descartes | 0.71 | 457.45 |
| POSTN | 0.0036784 | 184 | GTEx | DepMap | Descartes | 0.18 | 227.92 |
| PTPRG | 0.0036757 | 185 | GTEx | DepMap | Descartes | 0.39 | 165.28 |
| SVIL | 0.0036525 | 189 | GTEx | DepMap | Descartes | 0.20 | 95.98 |
| MEST | 0.0035499 | 204 | GTEx | DepMap | Descartes | 0.21 | 346.59 |
| LGALS1 | 0.0034835 | 215 | GTEx | DepMap | Descartes | 0.17 | 1135.55 |
| TMED9 | 0.0034512 | 220 | GTEx | DepMap | Descartes | 0.15 | 265.12 |
| MMP2 | 0.0028062 | 329 | GTEx | DepMap | Descartes | 0.12 | 143.11 |
| SSBP4 | 0.0027670 | 338 | GTEx | DepMap | Descartes | 0.24 | 528.94 |
| FZD2 | 0.0027124 | 343 | GTEx | DepMap | Descartes | 0.02 | 21.15 |
| PTBP1 | 0.0024729 | 406 | GTEx | DepMap | Descartes | 0.19 | 156.45 |
| TPBG | 0.0023069 | 453 | GTEx | DepMap | Descartes | 0.02 | 12.03 |
| SQSTM1 | 0.0022240 | 486 | GTEx | DepMap | Descartes | 0.19 | 248.04 |
| GDF15 | 0.0021861 | 498 | GTEx | DepMap | Descartes | 0.03 | 112.76 |
| NFIC | 0.0021669 | 504 | GTEx | DepMap | Descartes | 0.15 | 61.90 |
| PTPRK | 0.0021596 | 508 | GTEx | DepMap | Descartes | 0.39 | NA |
| CRTAP | 0.0021523 | 512 | GTEx | DepMap | Descartes | 0.06 | 37.51 |
| SPARC | 0.0021406 | 517 | GTEx | DepMap | Descartes | 0.27 | 314.80 |
| CALD1 | 0.0021119 | 533 | GTEx | DepMap | Descartes | 0.46 | 344.84 |
| SERPINH1 | 0.0020843 | 546 | GTEx | DepMap | Descartes | 0.05 | 58.12 |
| TPM2 | 0.0020322 | 569 | GTEx | DepMap | Descartes | 0.10 | 227.36 |
| STAT3 | 0.0019942 | 588 | GTEx | DepMap | Descartes | 0.20 | 153.21 |
Descartes adrenocortical markers
Top 50 marker genes of adrenocortical cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/adrenocortical/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 8.54e-01
Mean rank of genes in gene set: 6911.06
Median rank of genes in gene set: 8808.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| PDE10A | 0.0133388 | 10 | GTEx | DepMap | Descartes | 1.14 | 511.79 |
| IGF1R | 0.0038925 | 166 | GTEx | DepMap | Descartes | 0.53 | 170.96 |
| FREM2 | 0.0022173 | 489 | GTEx | DepMap | Descartes | 0.02 | 4.89 |
| FDXR | 0.0020164 | 578 | GTEx | DepMap | Descartes | 0.05 | 80.19 |
| DNER | 0.0018809 | 650 | GTEx | DepMap | Descartes | 0.10 | 137.20 |
| FDPS | 0.0017325 | 743 | GTEx | DepMap | Descartes | 0.17 | 318.82 |
| SCAP | 0.0017287 | 746 | GTEx | DepMap | Descartes | 0.12 | 106.38 |
| SCARB1 | 0.0017204 | 754 | GTEx | DepMap | Descartes | 0.09 | 63.76 |
| ERN1 | 0.0015148 | 924 | GTEx | DepMap | Descartes | 0.04 | 21.64 |
| JAKMIP2 | 0.0013120 | 1157 | GTEx | DepMap | Descartes | 0.20 | 87.49 |
| SH3PXD2B | 0.0013043 | 1167 | GTEx | DepMap | Descartes | 0.07 | 33.50 |
| GRAMD1B | 0.0008748 | 1849 | GTEx | DepMap | Descartes | 0.05 | 22.39 |
| INHA | 0.0004392 | 3388 | GTEx | DepMap | Descartes | 0.00 | 12.69 |
| STAR | 0.0000024 | 6420 | GTEx | DepMap | Descartes | 0.00 | 0.62 |
| GSTA4 | -0.0001058 | 7627 | GTEx | DepMap | Descartes | 0.07 | 165.40 |
| CYB5B | -0.0001151 | 7727 | GTEx | DepMap | Descartes | 0.06 | 60.55 |
| DHCR7 | -0.0001522 | 8140 | GTEx | DepMap | Descartes | 0.01 | 10.72 |
| FDX1 | -0.0002098 | 8725 | GTEx | DepMap | Descartes | 0.01 | 12.82 |
| TM7SF2 | -0.0002270 | 8892 | GTEx | DepMap | Descartes | 0.03 | 52.01 |
| BAIAP2L1 | -0.0002681 | 9244 | GTEx | DepMap | Descartes | 0.01 | 5.69 |
| NPC1 | -0.0002893 | 9405 | GTEx | DepMap | Descartes | 0.04 | 32.08 |
| HMGCR | -0.0003537 | 9866 | GTEx | DepMap | Descartes | 0.05 | 41.84 |
| DHCR24 | -0.0003579 | 9898 | GTEx | DepMap | Descartes | 0.03 | 18.56 |
| POR | -0.0003884 | 10093 | GTEx | DepMap | Descartes | 0.03 | 37.26 |
| LDLR | -0.0004603 | 10510 | GTEx | DepMap | Descartes | 0.01 | 6.69 |
| SLC16A9 | -0.0005036 | 10743 | GTEx | DepMap | Descartes | 0.03 | 22.74 |
| PAPSS2 | -0.0005714 | 10999 | GTEx | DepMap | Descartes | 0.01 | 8.93 |
| APOC1 | -0.0006880 | 11333 | GTEx | DepMap | Descartes | 0.04 | 212.45 |
| SH3BP5 | -0.0007278 | 11455 | GTEx | DepMap | Descartes | 0.02 | 27.29 |
| MSMO1 | -0.0008161 | 11632 | GTEx | DepMap | Descartes | 0.02 | 27.46 |
Descartes chromaffin markers
Top 50 marker genes of chromaffin cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/chromaffin/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 10955.71
Median rank of genes in gene set: 12399
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| RPH3A | 0.0031260 | 267 | GTEx | DepMap | Descartes | 0.05 | 37.81 |
| NTRK1 | 0.0010863 | 1460 | GTEx | DepMap | Descartes | 0.09 | 111.25 |
| CNKSR2 | 0.0006713 | 2452 | GTEx | DepMap | Descartes | 0.22 | 109.82 |
| ISL1 | 0.0003747 | 3722 | GTEx | DepMap | Descartes | 0.32 | 545.03 |
| GAL | 0.0001901 | 4920 | GTEx | DepMap | Descartes | 0.25 | 1249.44 |
| EYA1 | -0.0001206 | 7781 | GTEx | DepMap | Descartes | 0.30 | 288.57 |
| MAB21L1 | -0.0003386 | 9767 | GTEx | DepMap | Descartes | 0.04 | 56.88 |
| GREM1 | -0.0004890 | 10660 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MAB21L2 | -0.0008184 | 11637 | GTEx | DepMap | Descartes | 0.03 | 56.75 |
| SYNPO2 | -0.0010752 | 11979 | GTEx | DepMap | Descartes | 0.14 | 29.90 |
| MAP1B | -0.0011589 | 12048 | GTEx | DepMap | Descartes | 0.48 | 159.61 |
| CNTFR | -0.0012581 | 12113 | GTEx | DepMap | Descartes | 0.02 | 36.38 |
| PTCHD1 | -0.0013030 | 12139 | GTEx | DepMap | Descartes | 0.01 | 3.26 |
| REEP1 | -0.0013738 | 12203 | GTEx | DepMap | Descartes | 0.02 | 19.20 |
| SLC6A2 | -0.0014598 | 12241 | GTEx | DepMap | Descartes | 0.03 | 29.09 |
| RGMB | -0.0015034 | 12256 | GTEx | DepMap | Descartes | 0.05 | 45.82 |
| FAT3 | -0.0016154 | 12300 | GTEx | DepMap | Descartes | 0.01 | 1.46 |
| PLXNA4 | -0.0016767 | 12317 | GTEx | DepMap | Descartes | 0.02 | 5.02 |
| MLLT11 | -0.0017887 | 12352 | GTEx | DepMap | Descartes | 0.13 | 230.81 |
| TMEFF2 | -0.0018778 | 12368 | GTEx | DepMap | Descartes | 0.01 | 4.06 |
| ANKFN1 | -0.0020601 | 12399 | GTEx | DepMap | Descartes | 0.01 | 5.13 |
| HS3ST5 | -0.0020949 | 12408 | GTEx | DepMap | Descartes | 0.06 | 50.99 |
| TUBB2A | -0.0021334 | 12415 | GTEx | DepMap | Descartes | 0.05 | 114.00 |
| GAP43 | -0.0021974 | 12421 | GTEx | DepMap | Descartes | 0.06 | 119.66 |
| SLC44A5 | -0.0022902 | 12436 | GTEx | DepMap | Descartes | 0.03 | 13.58 |
| KCNB2 | -0.0023739 | 12446 | GTEx | DepMap | Descartes | 0.11 | 87.69 |
| ELAVL2 | -0.0025011 | 12463 | GTEx | DepMap | Descartes | 0.07 | 64.24 |
| BASP1 | -0.0025104 | 12465 | GTEx | DepMap | Descartes | 0.10 | 211.09 |
| EPHA6 | -0.0026143 | 12475 | GTEx | DepMap | Descartes | 0.06 | 20.47 |
| EYA4 | -0.0028666 | 12495 | GTEx | DepMap | Descartes | 0.07 | 43.35 |
Descartes Vascular_endothelial markers
Top 50 marker genes of Vascular_endothelial cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/vascular_endothelial/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 7.40e-01
Mean rank of genes in gene set: 6652.63
Median rank of genes in gene set: 6287
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| MYRIP | 0.0023291 | 446 | GTEx | DepMap | Descartes | 0.12 | 77.36 |
| PLVAP | 0.0012644 | 1209 | GTEx | DepMap | Descartes | 0.03 | 34.69 |
| HYAL2 | 0.0010028 | 1591 | GTEx | DepMap | Descartes | 0.04 | 37.86 |
| TMEM88 | 0.0007874 | 2083 | GTEx | DepMap | Descartes | 0.03 | 129.54 |
| SHANK3 | 0.0007856 | 2089 | GTEx | DepMap | Descartes | 0.03 | 14.17 |
| KDR | 0.0006637 | 2479 | GTEx | DepMap | Descartes | 0.01 | 6.03 |
| EHD3 | 0.0003916 | 3620 | GTEx | DepMap | Descartes | 0.01 | 9.66 |
| FLT4 | 0.0003355 | 3957 | GTEx | DepMap | Descartes | 0.01 | 5.09 |
| NOTCH4 | 0.0003089 | 4105 | GTEx | DepMap | Descartes | 0.02 | 7.71 |
| CDH5 | 0.0003075 | 4112 | GTEx | DepMap | Descartes | 0.01 | 7.00 |
| ESM1 | 0.0002447 | 4517 | GTEx | DepMap | Descartes | 0.00 | 7.46 |
| TIE1 | 0.0002101 | 4760 | GTEx | DepMap | Descartes | 0.01 | 7.26 |
| RASIP1 | 0.0002078 | 4785 | GTEx | DepMap | Descartes | 0.01 | 5.97 |
| ROBO4 | 0.0001939 | 4890 | GTEx | DepMap | Descartes | 0.01 | 4.75 |
| NPR1 | 0.0001784 | 4993 | GTEx | DepMap | Descartes | 0.00 | 2.18 |
| CLDN5 | 0.0001274 | 5365 | GTEx | DepMap | Descartes | 0.01 | 11.29 |
| MMRN2 | 0.0000905 | 5649 | GTEx | DepMap | Descartes | 0.00 | 4.78 |
| F8 | 0.0000662 | 5859 | GTEx | DepMap | Descartes | 0.00 | 0.98 |
| IRX3 | 0.0000312 | 6147 | GTEx | DepMap | Descartes | 0.00 | 3.98 |
| CRHBP | 0.0000017 | 6427 | GTEx | DepMap | Descartes | 0.00 | 1.72 |
| KANK3 | -0.0000390 | 6810 | GTEx | DepMap | Descartes | 0.00 | 4.76 |
| CALCRL | -0.0000767 | 7286 | GTEx | DepMap | Descartes | 0.01 | 7.43 |
| SLCO2A1 | -0.0001008 | 7561 | GTEx | DepMap | Descartes | 0.00 | 2.63 |
| PODXL | -0.0001166 | 7740 | GTEx | DepMap | Descartes | 0.02 | 14.25 |
| CYP26B1 | -0.0001320 | 7924 | GTEx | DepMap | Descartes | 0.00 | 1.35 |
| BTNL9 | -0.0001574 | 8205 | GTEx | DepMap | Descartes | 0.00 | 1.69 |
| GALNT15 | -0.0001660 | 8309 | GTEx | DepMap | Descartes | 0.00 | NA |
| RAMP2 | -0.0002445 | 9042 | GTEx | DepMap | Descartes | 0.02 | 87.78 |
| SHE | -0.0002577 | 9166 | GTEx | DepMap | Descartes | 0.00 | 0.76 |
| ID1 | -0.0004002 | 10172 | GTEx | DepMap | Descartes | 0.01 | 37.65 |
Descartes stromal markers
Top 50 marker genes of stromal cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/stromal/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.21e-01
Mean rank of genes in gene set: 5637.5
Median rank of genes in gene set: 6237.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ISLR | 0.0056121 | 75 | GTEx | DepMap | Descartes | 0.11 | 203.91 |
| COL1A1 | 0.0036897 | 182 | GTEx | DepMap | Descartes | 0.71 | 457.45 |
| POSTN | 0.0036784 | 184 | GTEx | DepMap | Descartes | 0.18 | 227.92 |
| CD248 | 0.0034068 | 225 | GTEx | DepMap | Descartes | 0.05 | 83.51 |
| COL1A2 | 0.0024567 | 412 | GTEx | DepMap | Descartes | 0.77 | 491.83 |
| PCOLCE | 0.0019153 | 627 | GTEx | DepMap | Descartes | 0.27 | 671.47 |
| COL3A1 | 0.0015482 | 893 | GTEx | DepMap | Descartes | 0.36 | 255.97 |
| COL6A3 | 0.0013348 | 1126 | GTEx | DepMap | Descartes | 0.11 | 40.36 |
| GLI2 | 0.0012038 | 1296 | GTEx | DepMap | Descartes | 0.02 | 10.13 |
| SFRP2 | 0.0011400 | 1389 | GTEx | DepMap | Descartes | 0.03 | 59.84 |
| ACTA2 | 0.0007822 | 2098 | GTEx | DepMap | Descartes | 0.05 | 142.89 |
| PRRX1 | 0.0007719 | 2122 | GTEx | DepMap | Descartes | 0.03 | 27.69 |
| PCDH18 | 0.0006914 | 2373 | GTEx | DepMap | Descartes | 0.01 | 4.32 |
| EDNRA | 0.0006047 | 2685 | GTEx | DepMap | Descartes | 0.03 | 19.48 |
| DCN | 0.0004397 | 3385 | GTEx | DepMap | Descartes | 0.04 | 19.63 |
| CDH11 | 0.0004155 | 3487 | GTEx | DepMap | Descartes | 0.06 | 30.69 |
| ELN | 0.0003291 | 3994 | GTEx | DepMap | Descartes | 0.05 | 49.62 |
| LUM | 0.0002989 | 4176 | GTEx | DepMap | Descartes | 0.05 | 64.58 |
| ABCC9 | 0.0001811 | 4979 | GTEx | DepMap | Descartes | 0.01 | 3.22 |
| LOX | 0.0001643 | 5113 | GTEx | DepMap | Descartes | 0.01 | 7.12 |
| ADAMTSL3 | 0.0000706 | 5824 | GTEx | DepMap | Descartes | 0.01 | 6.32 |
| COL12A1 | 0.0000571 | 5935 | GTEx | DepMap | Descartes | 0.03 | 10.04 |
| PDGFRA | -0.0000122 | 6540 | GTEx | DepMap | Descartes | 0.01 | 3.30 |
| DKK2 | -0.0000140 | 6556 | GTEx | DepMap | Descartes | 0.01 | 10.05 |
| COL27A1 | -0.0000253 | 6686 | GTEx | DepMap | Descartes | 0.02 | 5.38 |
| SCARA5 | -0.0000368 | 6788 | GTEx | DepMap | Descartes | 0.00 | 1.06 |
| ITGA11 | -0.0000436 | 6858 | GTEx | DepMap | Descartes | 0.01 | 2.39 |
| BICC1 | -0.0000636 | 7112 | GTEx | DepMap | Descartes | 0.10 | 63.39 |
| IGFBP3 | -0.0000761 | 7274 | GTEx | DepMap | Descartes | 0.01 | 16.09 |
| ADAMTS2 | -0.0000771 | 7287 | GTEx | DepMap | Descartes | 0.04 | 18.88 |
Descartes sympathoblasts markers
Top 50 marker genes of sympathoblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/sympathoblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 7.04e-01
Mean rank of genes in gene set: 6589.34
Median rank of genes in gene set: 9864.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CHGA | 0.0197936 | 1 | GTEx | DepMap | Descartes | 3.32 | 6803.85 |
| FAM155A | 0.0150706 | 7 | GTEx | DepMap | Descartes | 5.30 | 2348.91 |
| ST18 | 0.0130227 | 11 | GTEx | DepMap | Descartes | 0.51 | 354.01 |
| FGF14 | 0.0082252 | 36 | GTEx | DepMap | Descartes | 2.68 | 861.47 |
| SLC35F3 | 0.0079604 | 37 | GTEx | DepMap | Descartes | 0.34 | 397.51 |
| PCSK2 | 0.0075864 | 41 | GTEx | DepMap | Descartes | 0.41 | 351.78 |
| SLC18A1 | 0.0073638 | 45 | GTEx | DepMap | Descartes | 0.29 | 394.49 |
| CHGB | 0.0073304 | 46 | GTEx | DepMap | Descartes | 1.18 | 2045.75 |
| C1QL1 | 0.0054629 | 78 | GTEx | DepMap | Descartes | 0.17 | 485.19 |
| MGAT4C | 0.0034445 | 221 | GTEx | DepMap | Descartes | 0.28 | 43.18 |
| EML6 | 0.0031882 | 257 | GTEx | DepMap | Descartes | 0.42 | 169.87 |
| SLC24A2 | 0.0020566 | 559 | GTEx | DepMap | Descartes | 0.05 | 18.50 |
| ARC | 0.0015610 | 883 | GTEx | DepMap | Descartes | 0.07 | 101.27 |
| GCH1 | 0.0010517 | 1514 | GTEx | DepMap | Descartes | 0.14 | 200.77 |
| PACRG | 0.0009898 | 1616 | GTEx | DepMap | Descartes | 0.12 | 288.79 |
| DGKK | 0.0008425 | 1928 | GTEx | DepMap | Descartes | 0.01 | 5.59 |
| HTATSF1 | 0.0007916 | 2073 | GTEx | DepMap | Descartes | 0.09 | 124.61 |
| LAMA3 | 0.0003210 | 4041 | GTEx | DepMap | Descartes | 0.03 | 11.43 |
| PENK | -0.0002459 | 9058 | GTEx | DepMap | Descartes | 0.00 | 6.75 |
| SORCS3 | -0.0004909 | 10671 | GTEx | DepMap | Descartes | 0.00 | 1.95 |
| UNC80 | -0.0005340 | 10868 | GTEx | DepMap | Descartes | 0.18 | 52.23 |
| CNTN3 | -0.0005512 | 10924 | GTEx | DepMap | Descartes | 0.00 | 1.51 |
| KSR2 | -0.0009174 | 11802 | GTEx | DepMap | Descartes | 0.05 | 10.47 |
| CDH12 | -0.0010124 | 11920 | GTEx | DepMap | Descartes | 0.01 | 4.74 |
| TBX20 | -0.0011781 | 12064 | GTEx | DepMap | Descartes | 0.01 | 12.49 |
| TIAM1 | -0.0012614 | 12116 | GTEx | DepMap | Descartes | 0.14 | 64.04 |
| CCSER1 | -0.0013277 | 12157 | GTEx | DepMap | Descartes | 0.45 | NA |
| AGBL4 | -0.0013501 | 12178 | GTEx | DepMap | Descartes | 0.18 | 161.50 |
| ROBO1 | -0.0013647 | 12194 | GTEx | DepMap | Descartes | 0.41 | 168.16 |
| SPOCK3 | -0.0014473 | 12235 | GTEx | DepMap | Descartes | 0.03 | 45.09 |
Descartes erythroblasts markers
Top 50 marker genes of erythroblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/erythroblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.29e-01
Mean rank of genes in gene set: 7260.21
Median rank of genes in gene set: 7391
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| SELENBP1 | 0.0022309 | 483 | GTEx | DepMap | Descartes | 0.02 | 24.62 |
| DENND4A | 0.0011903 | 1317 | GTEx | DepMap | Descartes | 0.22 | 101.81 |
| SLC25A37 | 0.0010892 | 1456 | GTEx | DepMap | Descartes | 0.06 | 43.41 |
| ANK1 | 0.0009231 | 1742 | GTEx | DepMap | Descartes | 0.09 | 36.86 |
| TMCC2 | 0.0005295 | 2963 | GTEx | DepMap | Descartes | 0.02 | 17.70 |
| GCLC | 0.0002644 | 4387 | GTEx | DepMap | Descartes | 0.05 | 59.31 |
| CPOX | 0.0001947 | 4886 | GTEx | DepMap | Descartes | 0.01 | 21.02 |
| EPB41 | 0.0001717 | 5052 | GTEx | DepMap | Descartes | 0.13 | 76.35 |
| SLC4A1 | 0.0001447 | 5233 | GTEx | DepMap | Descartes | 0.00 | 1.09 |
| TRAK2 | 0.0000852 | 5692 | GTEx | DepMap | Descartes | 0.03 | 18.95 |
| XPO7 | 0.0000763 | 5768 | GTEx | DepMap | Descartes | 0.08 | 64.07 |
| SPECC1 | 0.0000112 | 6329 | GTEx | DepMap | Descartes | 0.01 | 3.18 |
| ALAS2 | -0.0000594 | 7061 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| BLVRB | -0.0000631 | 7106 | GTEx | DepMap | Descartes | 0.01 | 27.23 |
| MICAL2 | -0.0000867 | 7391 | GTEx | DepMap | Descartes | 0.02 | 7.15 |
| CAT | -0.0000969 | 7512 | GTEx | DepMap | Descartes | 0.02 | 30.16 |
| GYPC | -0.0001168 | 7743 | GTEx | DepMap | Descartes | 0.00 | 11.76 |
| RGS6 | -0.0001442 | 8040 | GTEx | DepMap | Descartes | 0.00 | 1.73 |
| SLC25A21 | -0.0002818 | 9348 | GTEx | DepMap | Descartes | 0.00 | 1.73 |
| MARCH3 | -0.0003302 | 9709 | GTEx | DepMap | Descartes | 0.04 | NA |
| SPTB | -0.0004136 | 10253 | GTEx | DepMap | Descartes | 0.01 | 2.83 |
| SNCA | -0.0004349 | 10389 | GTEx | DepMap | Descartes | 0.03 | 30.67 |
| RHD | -0.0004491 | 10460 | GTEx | DepMap | Descartes | 0.00 | 2.68 |
| FECH | -0.0004825 | 10627 | GTEx | DepMap | Descartes | 0.01 | 7.65 |
| RAPGEF2 | -0.0007503 | 11501 | GTEx | DepMap | Descartes | 0.13 | 65.29 |
| ABCB10 | -0.0008378 | 11673 | GTEx | DepMap | Descartes | 0.01 | 12.39 |
| TFR2 | -0.0008604 | 11714 | GTEx | DepMap | Descartes | 0.03 | 42.69 |
| SOX6 | -0.0015836 | 12287 | GTEx | DepMap | Descartes | 0.05 | 23.02 |
| TSPAN5 | -0.0022097 | 12424 | GTEx | DepMap | Descartes | 0.07 | 53.84 |
| NA | NA | NA | NA | NA | NA | NA | NA |
Descartes myeloid markers
Top 50 marker genes of myeloid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/myeloid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.91e-01
Mean rank of genes in gene set: 7664.53
Median rank of genes in gene set: 9065.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| SFMBT2 | 0.0031138 | 268 | GTEx | DepMap | Descartes | 0.16 | 72.48 |
| ITPR2 | 0.0023741 | 439 | GTEx | DepMap | Descartes | 0.13 | 41.72 |
| MARCH1 | 0.0022407 | 478 | GTEx | DepMap | Descartes | 0.11 | NA |
| CST3 | 0.0009987 | 1605 | GTEx | DepMap | Descartes | 0.10 | 124.78 |
| CTSB | 0.0008240 | 1995 | GTEx | DepMap | Descartes | 0.13 | 142.96 |
| PTPRE | 0.0008006 | 2051 | GTEx | DepMap | Descartes | 0.07 | 37.82 |
| RGL1 | 0.0005964 | 2707 | GTEx | DepMap | Descartes | 0.09 | 68.36 |
| LGMN | 0.0003120 | 4084 | GTEx | DepMap | Descartes | 0.04 | 69.27 |
| MERTK | 0.0002573 | 4440 | GTEx | DepMap | Descartes | 0.01 | 12.60 |
| TGFBI | 0.0001903 | 4918 | GTEx | DepMap | Descartes | 0.03 | 22.59 |
| CTSC | 0.0000767 | 5761 | GTEx | DepMap | Descartes | 0.02 | 12.73 |
| ADAP2 | 0.0000412 | 6061 | GTEx | DepMap | Descartes | 0.01 | 16.83 |
| CPVL | 0.0000393 | 6073 | GTEx | DepMap | Descartes | 0.02 | 33.56 |
| MSR1 | -0.0000007 | 6444 | GTEx | DepMap | Descartes | 0.03 | 30.60 |
| AXL | -0.0002007 | 8657 | GTEx | DepMap | Descartes | 0.01 | 12.00 |
| IFNGR1 | -0.0002055 | 8696 | GTEx | DepMap | Descartes | 0.04 | 59.95 |
| MS4A4A | -0.0002139 | 8770 | GTEx | DepMap | Descartes | 0.00 | 6.30 |
| CSF1R | -0.0002258 | 8872 | GTEx | DepMap | Descartes | 0.00 | 1.63 |
| SLC1A3 | -0.0002358 | 8977 | GTEx | DepMap | Descartes | 0.01 | 7.14 |
| CD163 | -0.0002557 | 9154 | GTEx | DepMap | Descartes | 0.00 | 1.96 |
| CTSD | -0.0002606 | 9187 | GTEx | DepMap | Descartes | 0.13 | 261.61 |
| ATP8B4 | -0.0002707 | 9261 | GTEx | DepMap | Descartes | 0.00 | 2.01 |
| CTSS | -0.0003105 | 9562 | GTEx | DepMap | Descartes | 0.01 | 7.61 |
| SLC9A9 | -0.0003191 | 9628 | GTEx | DepMap | Descartes | 0.02 | 26.47 |
| FGD2 | -0.0003362 | 9751 | GTEx | DepMap | Descartes | 0.00 | 0.32 |
| HRH1 | -0.0003407 | 9787 | GTEx | DepMap | Descartes | 0.01 | 5.14 |
| WWP1 | -0.0003650 | 9941 | GTEx | DepMap | Descartes | 0.04 | 28.69 |
| SLCO2B1 | -0.0003824 | 10056 | GTEx | DepMap | Descartes | 0.01 | 2.87 |
| CYBB | -0.0004160 | 10268 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| FGL2 | -0.0004258 | 10326 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
Descartes Schwann markers
Top 50 marker genes of Schwann cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/schwann/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.96e-01
Mean rank of genes in gene set: 7742.36
Median rank of genes in gene set: 8079.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ERBB4 | 0.0192517 | 2 | GTEx | DepMap | Descartes | 4.42 | 1593.77 |
| NRXN3 | 0.0033081 | 238 | GTEx | DepMap | Descartes | 0.24 | 110.40 |
| STARD13 | 0.0017572 | 733 | GTEx | DepMap | Descartes | 0.09 | 54.10 |
| PTN | 0.0016322 | 817 | GTEx | DepMap | Descartes | 0.13 | 350.67 |
| LAMC1 | 0.0014763 | 961 | GTEx | DepMap | Descartes | 0.11 | 51.37 |
| DST | 0.0011894 | 1319 | GTEx | DepMap | Descartes | 0.76 | 138.66 |
| FIGN | 0.0010572 | 1505 | GTEx | DepMap | Descartes | 0.10 | 42.72 |
| PLCE1 | 0.0008043 | 2044 | GTEx | DepMap | Descartes | 0.05 | 12.29 |
| COL5A2 | 0.0005867 | 2742 | GTEx | DepMap | Descartes | 0.10 | 53.22 |
| SCN7A | 0.0005567 | 2856 | GTEx | DepMap | Descartes | 0.07 | 42.18 |
| OLFML2A | 0.0003571 | 3827 | GTEx | DepMap | Descartes | 0.01 | 3.47 |
| COL18A1 | 0.0001872 | 4940 | GTEx | DepMap | Descartes | 0.09 | 48.00 |
| MPZ | 0.0001079 | 5512 | GTEx | DepMap | Descartes | 0.00 | 11.43 |
| ADAMTS5 | 0.0000805 | 5733 | GTEx | DepMap | Descartes | 0.01 | 1.81 |
| SFRP1 | -0.0000466 | 6893 | GTEx | DepMap | Descartes | 0.04 | 32.06 |
| EDNRB | -0.0000476 | 6907 | GTEx | DepMap | Descartes | 0.00 | 1.25 |
| HMGA2 | -0.0000612 | 7078 | GTEx | DepMap | Descartes | 0.00 | 1.06 |
| GAS7 | -0.0000806 | 7327 | GTEx | DepMap | Descartes | 0.02 | 11.08 |
| GRIK3 | -0.0000883 | 7412 | GTEx | DepMap | Descartes | 0.01 | 2.17 |
| LAMA4 | -0.0001188 | 7763 | GTEx | DepMap | Descartes | 0.02 | 8.48 |
| NRXN1 | -0.0001336 | 7932 | GTEx | DepMap | Descartes | 0.48 | 195.21 |
| MDGA2 | -0.0001357 | 7956 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ERBB3 | -0.0001572 | 8203 | GTEx | DepMap | Descartes | 0.00 | 2.78 |
| PLP1 | -0.0002035 | 8683 | GTEx | DepMap | Descartes | 0.00 | 0.47 |
| PTPRZ1 | -0.0003227 | 9656 | GTEx | DepMap | Descartes | 0.00 | 0.73 |
| VIM | -0.0003230 | 9659 | GTEx | DepMap | Descartes | 0.14 | 210.72 |
| LAMB1 | -0.0004220 | 10299 | GTEx | DepMap | Descartes | 0.12 | 79.80 |
| GFRA3 | -0.0004861 | 10649 | GTEx | DepMap | Descartes | 0.03 | 58.24 |
| COL25A1 | -0.0005436 | 10900 | GTEx | DepMap | Descartes | 0.00 | 0.53 |
| SLC35F1 | -0.0006305 | 11186 | GTEx | DepMap | Descartes | 0.01 | 5.16 |
Descartes Megakaryocytes markers
Top 50 marker genes of Megakaryocytes cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/megakaryocytes/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 6.10e-01
Mean rank of genes in gene set: 6425.07
Median rank of genes in gene set: 6539
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| MCTP1 | 0.0050385 | 87 | GTEx | DepMap | Descartes | 0.20 | 162.21 |
| FLNA | 0.0043616 | 134 | GTEx | DepMap | Descartes | 0.26 | 120.95 |
| MYH9 | 0.0027806 | 336 | GTEx | DepMap | Descartes | 0.14 | 77.11 |
| TPM4 | 0.0023819 | 437 | GTEx | DepMap | Descartes | 0.17 | 133.13 |
| VCL | 0.0015096 | 927 | GTEx | DepMap | Descartes | 0.08 | 38.60 |
| LIMS1 | 0.0010556 | 1507 | GTEx | DepMap | Descartes | 0.11 | 104.67 |
| ACTB | 0.0010535 | 1511 | GTEx | DepMap | Descartes | 1.52 | 2886.06 |
| TUBB1 | 0.0010009 | 1596 | GTEx | DepMap | Descartes | 0.00 | 1.97 |
| ANGPT1 | 0.0009883 | 1618 | GTEx | DepMap | Descartes | 0.02 | 17.44 |
| ZYX | 0.0009425 | 1704 | GTEx | DepMap | Descartes | 0.06 | 106.24 |
| TGFB1 | 0.0008181 | 2013 | GTEx | DepMap | Descartes | 0.07 | 102.79 |
| GSN | 0.0006837 | 2399 | GTEx | DepMap | Descartes | 0.06 | 37.35 |
| MED12L | 0.0006739 | 2441 | GTEx | DepMap | Descartes | 0.08 | 28.51 |
| GP1BA | 0.0006074 | 2676 | GTEx | DepMap | Descartes | 0.01 | 16.64 |
| STON2 | 0.0005014 | 3090 | GTEx | DepMap | Descartes | 0.04 | 37.18 |
| P2RX1 | 0.0003265 | 4012 | GTEx | DepMap | Descartes | 0.00 | 3.60 |
| ITGA2B | 0.0002921 | 4215 | GTEx | DepMap | Descartes | 0.02 | 18.59 |
| SPN | 0.0002694 | 4352 | GTEx | DepMap | Descartes | 0.01 | 6.34 |
| PSTPIP2 | 0.0002457 | 4510 | GTEx | DepMap | Descartes | 0.01 | 10.39 |
| TLN1 | 0.0002235 | 4663 | GTEx | DepMap | Descartes | 0.03 | 14.24 |
| LTBP1 | 0.0000791 | 5741 | GTEx | DepMap | Descartes | 0.02 | 10.59 |
| RAP1B | 0.0000213 | 6237 | GTEx | DepMap | Descartes | 0.06 | 19.60 |
| ARHGAP6 | -0.0000122 | 6539 | GTEx | DepMap | Descartes | 0.01 | 6.40 |
| FERMT3 | -0.0000292 | 6716 | GTEx | DepMap | Descartes | 0.01 | 11.13 |
| ITGB3 | -0.0001018 | 7572 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| STOM | -0.0001163 | 7736 | GTEx | DepMap | Descartes | 0.01 | 12.88 |
| TRPC6 | -0.0001396 | 7998 | GTEx | DepMap | Descartes | 0.00 | 2.08 |
| MYLK | -0.0001620 | 8257 | GTEx | DepMap | Descartes | 0.01 | 2.75 |
| FLI1 | -0.0002034 | 8682 | GTEx | DepMap | Descartes | 0.02 | 11.80 |
| CD84 | -0.0002278 | 8904 | GTEx | DepMap | Descartes | 0.00 | 2.07 |
Descartes Lyphoid markers
Top 50 marker genes of Lyphoid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/lymphoid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.96e-01
Mean rank of genes in gene set: 7764.83
Median rank of genes in gene set: 9852
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| RAP1GAP2 | 0.0079040 | 38 | GTEx | DepMap | Descartes | 0.92 | 506.60 |
| TOX | 0.0028182 | 326 | GTEx | DepMap | Descartes | 0.64 | 662.76 |
| ARID5B | 0.0017972 | 707 | GTEx | DepMap | Descartes | 0.09 | 46.54 |
| FOXP1 | 0.0016848 | 784 | GTEx | DepMap | Descartes | 0.56 | 239.50 |
| PLEKHA2 | 0.0013744 | 1082 | GTEx | DepMap | Descartes | 0.04 | 30.41 |
| IKZF1 | 0.0012076 | 1287 | GTEx | DepMap | Descartes | 0.02 | 16.82 |
| MBNL1 | 0.0010852 | 1463 | GTEx | DepMap | Descartes | 0.22 | 148.24 |
| MCTP2 | 0.0010667 | 1489 | GTEx | DepMap | Descartes | 0.02 | 10.66 |
| ETS1 | 0.0008682 | 1864 | GTEx | DepMap | Descartes | 0.05 | 34.66 |
| CELF2 | 0.0008241 | 1994 | GTEx | DepMap | Descartes | 0.10 | 50.27 |
| WIPF1 | 0.0004944 | 3118 | GTEx | DepMap | Descartes | 0.10 | 80.16 |
| CCND3 | 0.0002806 | 4291 | GTEx | DepMap | Descartes | 0.05 | 78.27 |
| NCALD | 0.0001990 | 4860 | GTEx | DepMap | Descartes | 0.07 | 69.28 |
| ITPKB | 0.0000899 | 5655 | GTEx | DepMap | Descartes | 0.01 | 7.39 |
| LCP1 | 0.0000577 | 5926 | GTEx | DepMap | Descartes | 0.01 | 12.81 |
| ABLIM1 | -0.0000365 | 6783 | GTEx | DepMap | Descartes | 0.08 | 35.90 |
| MSN | -0.0002122 | 8755 | GTEx | DepMap | Descartes | 0.02 | 19.77 |
| ARHGDIB | -0.0002333 | 8953 | GTEx | DepMap | Descartes | 0.00 | 14.47 |
| B2M | -0.0002468 | 9069 | GTEx | DepMap | Descartes | 0.32 | 531.16 |
| PITPNC1 | -0.0002850 | 9373 | GTEx | DepMap | Descartes | 0.22 | 121.96 |
| GNG2 | -0.0003411 | 9791 | GTEx | DepMap | Descartes | 0.15 | 151.93 |
| RCSD1 | -0.0003605 | 9913 | GTEx | DepMap | Descartes | 0.00 | 2.19 |
| CCL5 | -0.0003680 | 9955 | GTEx | DepMap | Descartes | 0.00 | 3.81 |
| DOCK10 | -0.0003756 | 10011 | GTEx | DepMap | Descartes | 0.10 | 57.16 |
| LEF1 | -0.0003787 | 10031 | GTEx | DepMap | Descartes | 0.00 | 2.33 |
| SAMD3 | -0.0003819 | 10052 | GTEx | DepMap | Descartes | 0.00 | 1.62 |
| PRKCH | -0.0004751 | 10589 | GTEx | DepMap | Descartes | 0.02 | 15.86 |
| ANKRD44 | -0.0005144 | 10790 | GTEx | DepMap | Descartes | 0.08 | 44.81 |
| SP100 | -0.0005948 | 11079 | GTEx | DepMap | Descartes | 0.01 | 7.52 |
| EVL | -0.0006165 | 11145 | GTEx | DepMap | Descartes | 0.27 | 265.69 |
Below shows the ranks on this GEP for immune marker gene lists curated by CellTypist (https://www.celltypist.org/encyclopedia).
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Fibroblasts: Fibroblasts (curated markers)
the most common cells of connective tissues which synthesize the extracellular matrix and collagen to maintain tissue homeostasis:
Wilcoxon ranksum test P-value for gene set overrepresentation: 8.99e-03
Mean rank of genes in gene set: 1326.33
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| COL1A1 | 0.0036897 | 182 | GTEx | DepMap | Descartes | 0.71 | 457.45 |
| COL1A2 | 0.0024567 | 412 | GTEx | DepMap | Descartes | 0.77 | 491.83 |
| DCN | 0.0004397 | 3385 | GTEx | DepMap | Descartes | 0.04 | 19.63 |
Erythroid: Mid erythroid (curated markers)
middle erythroid cells in fetal liver:
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.37e-02
Mean rank of genes in gene set: 1197
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| KCNH2 | 0.0043547 | 135 | GTEx | DepMap | Descartes | 0.30 | 274.48 |
| PRDX2 | 0.0007288 | 2259 | GTEx | DepMap | Descartes | 0.24 | 584.69 |
ILC: ILC (curated markers)
specialised innate immune cells from the lymphoid lineage but without antigen-specific T-cell receptors on the surface:
Wilcoxon ranksum test P-value for gene set overrepresentation: 3.29e-02
Mean rank of genes in gene set: 2426
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TLE1 | 0.0043539 | 136 | GTEx | DepMap | Descartes | 0.18 | 188.54 |
| S100A13 | 0.0032732 | 245 | GTEx | DepMap | Descartes | 0.07 | 126.44 |
| AREG | -0.0000470 | 6897 | GTEx | DepMap | Descartes | 0.00 | 0.00 |