Program: 24. Peripheral Nervous System III.
Submit a comment on this gene expression program’s interpretation: CLICK
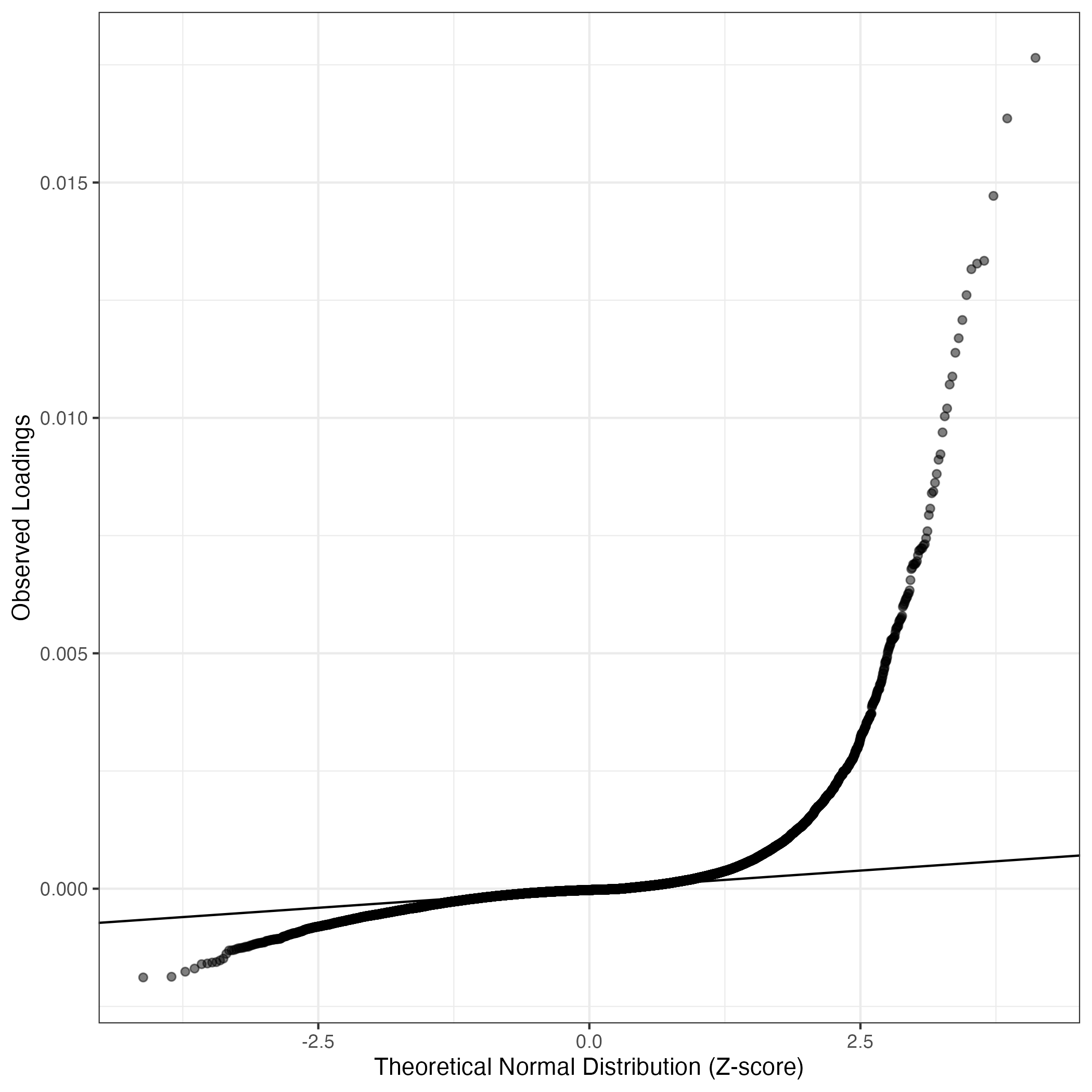
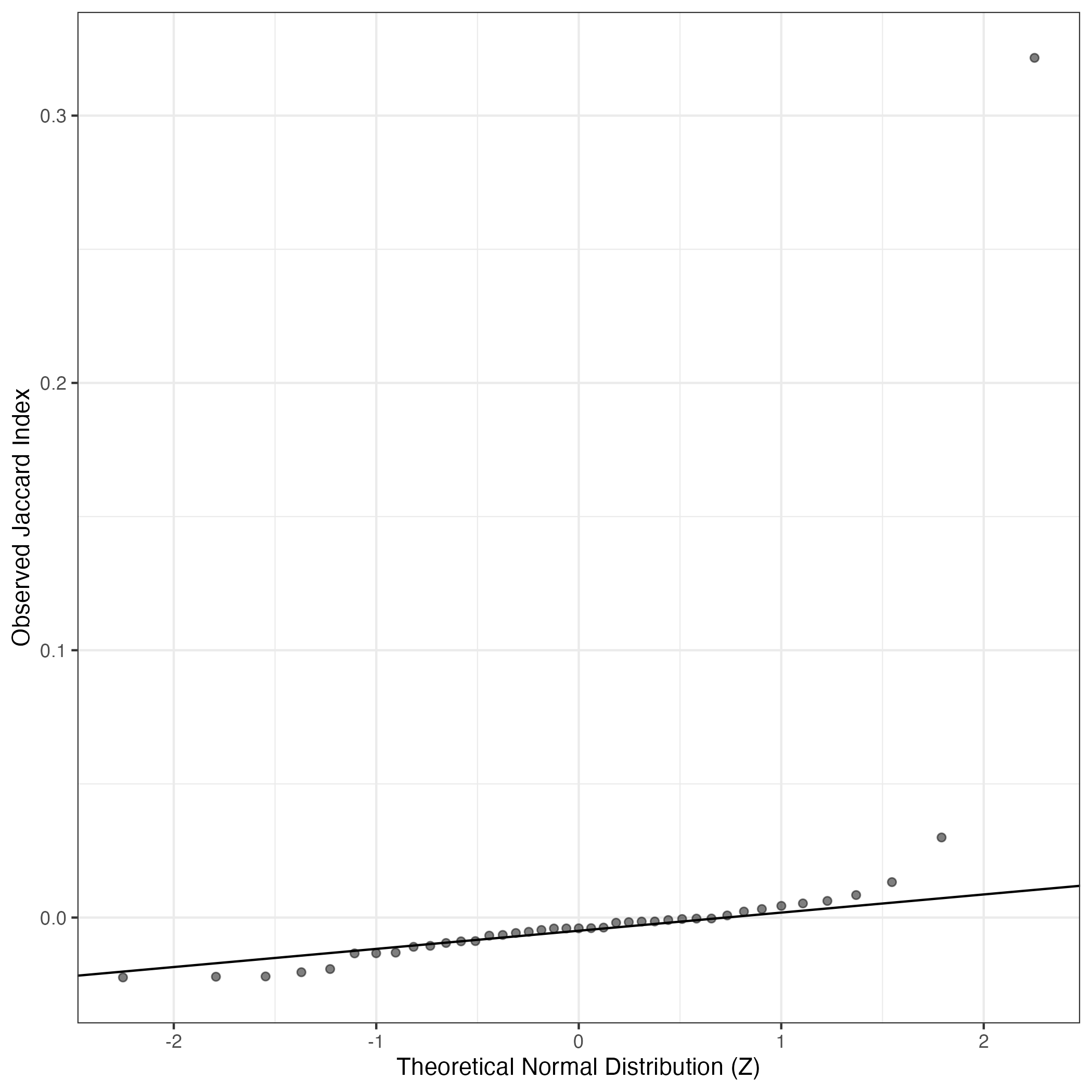
QQ-plot of gene loadings, averaged over both independent splits of the data
This plot highlights the relative contribution of each gene to the GEP
Top genes driving this program.
Note: Decartes website is buggy, try refreshing. Also, Decartes fetal adrenal data have been collected at specific time points (89-122 days), all possible cell types of interest may not be represented, do not overinterpret.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
| Gene | Loading | Gene.Name | GTEx | DepMap | Descartes | Mean.Counts | Mean.Tpm | |
|---|---|---|---|---|---|---|---|---|
| 1 | CHGA | 0.0176475 | chromogranin A | GTEx | DepMap | Descartes | 94.32 | 11651.42 |
| 2 | CHGB | 0.0163608 | chromogranin B | GTEx | DepMap | Descartes | 49.98 | 4972.72 |
| 3 | SCG2 | 0.0147153 | secretogranin II | GTEx | DepMap | Descartes | 12.62 | 1285.25 |
| 4 | RAMP1 | 0.0133373 | receptor activity modifying protein 1 | GTEx | DepMap | Descartes | 8.76 | 2334.58 |
| 5 | PENK | 0.0132775 | proenkephalin | GTEx | DepMap | Descartes | 8.46 | 1571.03 |
| 6 | INSM1 | 0.0131589 | INSM transcriptional repressor 1 | GTEx | DepMap | Descartes | 3.54 | 307.92 |
| 7 | C1QL1 | 0.0126086 | complement C1q like 1 | GTEx | DepMap | Descartes | 4.41 | 675.65 |
| 8 | TH | 0.0120792 | tyrosine hydroxylase | GTEx | DepMap | Descartes | 10.05 | 1450.43 |
| 9 | CARTPT | 0.0116929 | CART prepropeptide | GTEx | DepMap | Descartes | 7.50 | 1867.42 |
| 10 | DBH | 0.0113835 | dopamine beta-hydroxylase | GTEx | DepMap | Descartes | 11.94 | 1169.41 |
| 11 | HTATSF1 | 0.0108788 | HIV-1 Tat specific factor 1 | GTEx | DepMap | Descartes | 8.81 | 746.65 |
| 12 | SLC18A1 | 0.0107091 | solute carrier family 18 member A1 | GTEx | DepMap | Descartes | 3.69 | 355.15 |
| 13 | PCSK1N | 0.0102016 | proprotein convertase subtilisin/kexin type 1 inhibitor | GTEx | DepMap | Descartes | 14.78 | 3168.28 |
| 14 | PNMT | 0.0100336 | phenylethanolamine N-methyltransferase | GTEx | DepMap | Descartes | 6.62 | 1387.62 |
| 15 | ARC | 0.0096922 | activity regulated cytoskeleton associated protein | GTEx | DepMap | Descartes | 2.95 | 224.41 |
| 16 | CYB561 | 0.0092274 | cytochrome b561 | GTEx | DepMap | Descartes | 3.01 | 267.13 |
| 17 | CHRNA3 | 0.0091114 | cholinergic receptor nicotinic alpha 3 subunit | GTEx | DepMap | Descartes | 3.53 | 326.62 |
| 18 | NT5DC2 | 0.0088095 | 5’-nucleotidase domain containing 2 | GTEx | DepMap | Descartes | 7.14 | 867.51 |
| 19 | SLC24A2 | 0.0086225 | solute carrier family 24 member 2 | GTEx | DepMap | Descartes | 1.25 | 33.30 |
| 20 | VWA5B2 | 0.0084371 | von Willebrand factor A domain containing 5B2 | GTEx | DepMap | Descartes | 0.93 | 61.06 |
| 21 | FAM162B | 0.0084005 | family with sequence similarity 162 member B | GTEx | DepMap | Descartes | 2.35 | 508.56 |
| 22 | SERTM2 | 0.0080740 | serine rich and transmembrane domain containing 2 | GTEx | DepMap | Descartes | 0.78 | NA |
| 23 | MIR7-3HG | 0.0079361 | MIR7-3 host gene | GTEx | DepMap | Descartes | 0.73 | 104.42 |
| 24 | QDPR | 0.0075939 | quinoid dihydropteridine reductase | GTEx | DepMap | Descartes | 4.46 | 649.01 |
| 25 | RGS4 | 0.0074418 | regulator of G protein signaling 4 | GTEx | DepMap | Descartes | 7.25 | 516.11 |
| 26 | SCG5 | 0.0073173 | secretogranin V | GTEx | DepMap | Descartes | 3.46 | 602.98 |
| 27 | ARFGEF3 | 0.0072925 | ARFGEF family member 3 | GTEx | DepMap | Descartes | 1.13 | NA |
| 28 | CNTN1 | 0.0072304 | contactin 1 | GTEx | DepMap | Descartes | 4.43 | 209.12 |
| 29 | PCSK2 | 0.0072290 | proprotein convertase subtilisin/kexin type 2 | GTEx | DepMap | Descartes | 1.40 | 82.06 |
| 30 | SLC35D3 | 0.0071926 | solute carrier family 35 member D3 | GTEx | DepMap | Descartes | 0.38 | 33.13 |
| 31 | ANKRD29 | 0.0071797 | ankyrin repeat domain 29 | GTEx | DepMap | Descartes | 1.29 | 92.65 |
| 32 | MEG3 | 0.0070788 | maternally expressed 3 | GTEx | DepMap | Descartes | 69.04 | 1877.73 |
| 33 | DGKK | 0.0069569 | diacylglycerol kinase kappa | GTEx | DepMap | Descartes | 1.07 | 37.75 |
| 34 | SEZ6L2 | 0.0069160 | seizure related 6 homolog like 2 | GTEx | DepMap | Descartes | 2.79 | 199.95 |
| 35 | RGS5 | 0.0068912 | regulator of G protein signaling 5 | GTEx | DepMap | Descartes | 15.06 | 667.31 |
| 36 | VGF | 0.0068901 | VGF nerve growth factor inducible | GTEx | DepMap | Descartes | 1.57 | 138.81 |
| 37 | CCSER1 | 0.0068894 | coiled-coil serine rich protein 1 | GTEx | DepMap | Descartes | 1.38 | NA |
| 38 | ST18 | 0.0068163 | ST18 C2H2C-type zinc finger transcription factor | GTEx | DepMap | Descartes | 0.94 | 46.16 |
| 39 | CNIH2 | 0.0067892 | cornichon family AMPA receptor auxiliary protein 2 | GTEx | DepMap | Descartes | 1.07 | 153.57 |
| 40 | DLK1 | 0.0065554 | delta like non-canonical Notch ligand 1 | GTEx | DepMap | Descartes | 46.63 | 2296.78 |
| 41 | CFAP61 | 0.0063367 | cilia and flagella associated protein 61 | GTEx | DepMap | Descartes | 0.40 | NA |
| 42 | NLRP1 | 0.0062756 | NLR family pyrin domain containing 1 | GTEx | DepMap | Descartes | 2.75 | 110.96 |
| 43 | P4HTM | 0.0062610 | prolyl 4-hydroxylase, transmembrane | GTEx | DepMap | Descartes | 1.86 | 82.01 |
| 44 | C2CD4A | 0.0062009 | C2 calcium dependent domain containing 4A | GTEx | DepMap | Descartes | 0.50 | 40.26 |
| 45 | SYT4 | 0.0061707 | synaptotagmin 4 | GTEx | DepMap | Descartes | 1.21 | 80.02 |
| 46 | ACE | 0.0061431 | angiotensin I converting enzyme | GTEx | DepMap | Descartes | 0.69 | 37.25 |
| 47 | ZDBF2 | 0.0060973 | zinc finger DBF-type containing 2 | GTEx | DepMap | Descartes | 2.43 | 71.71 |
| 48 | HACD3 | 0.0060495 | 3-hydroxyacyl-CoA dehydratase 3 | GTEx | DepMap | Descartes | 4.92 | NA |
| 49 | SLC16A12 | 0.0060204 | solute carrier family 16 member 12 | GTEx | DepMap | Descartes | 0.60 | 37.58 |
| 50 | UNC5A | 0.0059863 | unc-5 netrin receptor A | GTEx | DepMap | Descartes | 0.73 | 53.30 |
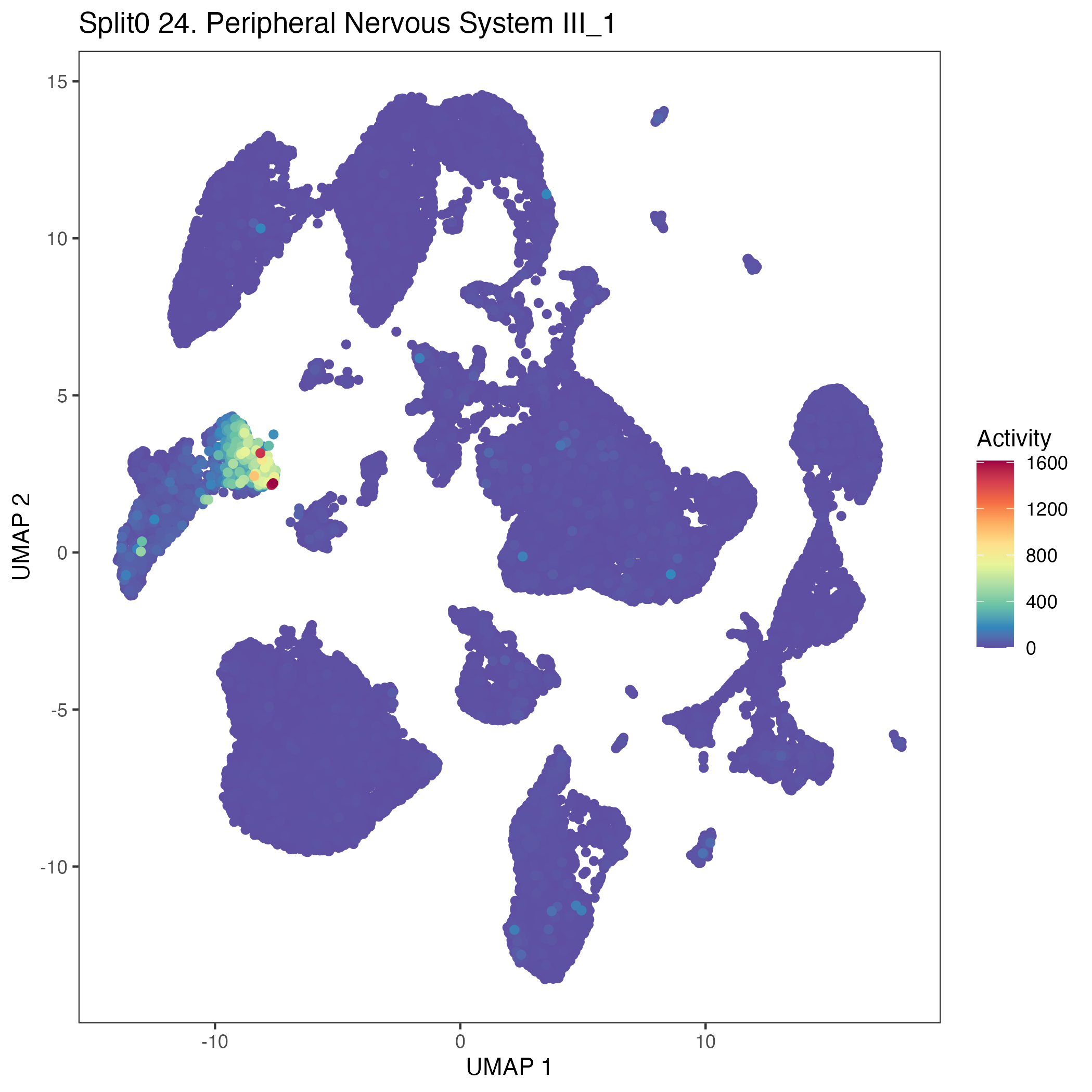
UMAP plots showing activity of gene expression program identified in GEP 24. Peripheral Nervous System III:
Gene set Enrichments for this program, caculated from top 50 genes
mSigDB Cell Types Gene Set:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| DESCARTES_MAIN_FETAL_SYMPATHOBLASTS | 3.26e-39 | 411.99 | 202.40 | 2.19e-36 | 2.19e-36 | 19CHGA, CHGB, SCG2, PENK, C1QL1, TH, CARTPT, DBH, HTATSF1, SLC18A1, PNMT, ARC, CYB561, NT5DC2, SERTM2, QDPR, NLRP1, ZDBF2, HACD3 |
57 |
| BUSSLINGER_GASTRIC_ANTRAL_ECS | 6.08e-09 | 278.88 | 59.22 | 2.15e-07 | 4.08e-06 | 4SCG2, PCSK1N, SCG5, MEG3 |
12 |
| DESCARTES_FETAL_ADRENAL_SYMPATHOBLASTS | 5.32e-22 | 97.30 | 46.85 | 1.78e-19 | 3.57e-19 | 14CHGA, PENK, CARTPT, PNMT, ARC, SLC24A2, VWA5B2, SERTM2, MIR7-3HG, PCSK2, VGF, CCSER1, ST18, C2CD4A |
117 |
| BUSSLINGER_GASTRIC_D_CELLS | 2.10e-09 | 124.04 | 35.28 | 9.39e-08 | 1.41e-06 | 5CHGB, PCSK1N, SCG5, ARFGEF3, PCSK2 |
28 |
| BUSSLINGER_DUODENAL_EC_CELLS | 3.61e-09 | 109.69 | 31.48 | 1.35e-07 | 2.42e-06 | 5CHGA, CHGB, SCG2, PCSK1N, PCSK2 |
31 |
| BUSSLINGER_GASTRIC_OXYNTIC_ENTEROCHROMAFFIN_LIKE_CELLS | 6.42e-14 | 74.19 | 30.66 | 7.18e-12 | 4.31e-11 | 9CHGA, CHGB, SCG2, PCSK1N, QDPR, SCG5, ARFGEF3, SEZ6L2, VGF |
85 |
| DESCARTES_FETAL_HEART_VISCERAL_NEURONS | 5.58e-12 | 62.03 | 24.37 | 4.16e-10 | 3.75e-09 | 8CHGB, SCG2, TH, DBH, SLC18A1, CHRNA3, RGS4, VGF |
87 |
| TRAVAGLINI_LUNG_NEUROENDOCRINE_CELL | 5.20e-14 | 54.49 | 23.73 | 6.98e-12 | 3.49e-11 | 10CHGB, SCG2, INSM1, PCSK1N, MIR7-3HG, SCG5, PCSK2, SLC35D3, MEG3, SYT4 |
128 |
| BUSSLINGER_DUODENAL_I_CELLS | 3.76e-06 | 126.05 | 22.32 | 1.01e-04 | 2.52e-03 | 3SCG2, PCSK1N, SCG5 |
16 |
| DESCARTES_FETAL_LUNG_NEUROENDOCRINE_CELLS | 3.94e-10 | 51.05 | 18.83 | 2.03e-08 | 2.64e-07 | 7CHGA, INSM1, SLC18A1, VWA5B2, RGS4, VGF, SLC16A12 |
89 |
| DESCARTES_FETAL_STOMACH_NEUROENDOCRINE_CELLS | 4.36e-08 | 63.50 | 18.80 | 1.46e-06 | 2.93e-05 | 5CHGA, SLC18A1, VWA5B2, RGS4, ST18 |
50 |
| GAO_LARGE_INTESTINE_ADULT_CA_ENTEROENDOCRINE_CELLS | 1.58e-17 | 37.16 | 18.65 | 2.66e-15 | 1.06e-14 | 15CHGB, SCG2, RAMP1, INSM1, SLC18A1, PCSK1N, ARC, VWA5B2, RGS4, SCG5, VGF, ST18, CNIH2, SYT4, SLC16A12 |
312 |
| DESCARTES_FETAL_INTESTINE_CHROMAFFIN_CELLS | 4.35e-11 | 47.07 | 18.65 | 2.92e-09 | 2.92e-08 | 8CHGA, SCG2, SLC18A1, PCSK1N, PNMT, VWA5B2, MIR7-3HG, ST18 |
112 |
| BUSSLINGER_DUODENAL_K_CELLS | 7.60e-06 | 96.27 | 17.52 | 1.82e-04 | 5.10e-03 | 3CHGA, SCG2, SCG5 |
20 |
| BUSSLINGER_GASTRIC_G_CELLS | 7.79e-10 | 45.99 | 17.02 | 3.73e-08 | 5.23e-07 | 7CHGB, SCG2, INSM1, PCSK1N, SCG5, ARFGEF3, PCSK2 |
98 |
| MURARO_PANCREAS_ALPHA_CELL | 7.23e-18 | 26.35 | 13.83 | 1.62e-15 | 4.85e-15 | 18CHGA, CHGB, SCG2, PCSK1N, VWA5B2, QDPR, RGS4, SCG5, ARFGEF3, CNTN1, PCSK2, SEZ6L2, VGF, ST18, CNIH2, P4HTM, C2CD4A, SYT4 |
568 |
| ZHONG_PFC_C2_THY1_POS_OPC | 1.93e-05 | 68.27 | 12.75 | 4.29e-04 | 1.30e-02 | 3PCSK1N, SCG5, CNTN1 |
27 |
| BUSSLINGER_GASTRIC_X_CELLS | 3.02e-09 | 26.78 | 10.72 | 1.19e-07 | 2.02e-06 | 8CHGB, PCSK1N, VWA5B2, QDPR, ARFGEF3, SEZ6L2, ST18, SYT4 |
191 |
| MANNO_MIDBRAIN_NEUROTYPES_HSERT | 1.84e-12 | 20.70 | 10.03 | 1.55e-10 | 1.24e-09 | 13CHGA, CHGB, SCG2, PCSK1N, QDPR, SCG5, ARFGEF3, CNTN1, MEG3, SEZ6L2, VGF, CCSER1, CNIH2 |
450 |
| MANNO_MIDBRAIN_NEUROTYPES_HDA1 | 1.38e-13 | 19.41 | 9.79 | 1.32e-11 | 9.23e-11 | 15CHGA, SCG2, INSM1, TH, SLC18A1, PCSK1N, SCG5, CNTN1, PCSK2, MEG3, SEZ6L2, VGF, CCSER1, DLK1, SYT4 |
584 |
Dowload full table
mSigDB Hallmark Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| HALLMARK_PANCREAS_BETA_CELLS | 6.40e-05 | 44.36 | 8.46 | 3.20e-03 | 3.20e-03 | 3CHGA, INSM1, PCSK2 |
40 |
| HALLMARK_SPERMATOGENESIS | 1.34e-04 | 17.09 | 4.41 | 3.35e-03 | 6.70e-03 | 4PCSK1N, SCG5, CNIH2, ACE |
135 |
| HALLMARK_KRAS_SIGNALING_UP | 6.73e-03 | 8.34 | 1.65 | 1.12e-01 | 3.37e-01 | 3PCSK1N, SCG5, ACE |
200 |
| HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | 5.67e-02 | 5.42 | 0.63 | 6.74e-01 | 1.00e+00 | 2SCG2, RGS4 |
200 |
| HALLMARK_HEDGEHOG_SIGNALING | 6.74e-02 | 15.01 | 0.36 | 6.74e-01 | 1.00e+00 | 1SCG2 |
36 |
| HALLMARK_UV_RESPONSE_DN | 2.43e-01 | 3.68 | 0.09 | 1.00e+00 | 1.00e+00 | 1RGS4 |
144 |
| HALLMARK_IL2_STAT5_SIGNALING | 3.19e-01 | 2.66 | 0.07 | 1.00e+00 | 1.00e+00 | 1PENK |
199 |
| HALLMARK_ADIPOGENESIS | 3.20e-01 | 2.64 | 0.07 | 1.00e+00 | 1.00e+00 | 1QDPR |
200 |
| HALLMARK_ESTROGEN_RESPONSE_LATE | 3.20e-01 | 2.64 | 0.07 | 1.00e+00 | 1.00e+00 | 1TH |
200 |
| HALLMARK_APICAL_JUNCTION | 3.20e-01 | 2.64 | 0.07 | 1.00e+00 | 1.00e+00 | 1CNTN1 |
200 |
| HALLMARK_MTORC1_SIGNALING | 3.20e-01 | 2.64 | 0.07 | 1.00e+00 | 1.00e+00 | 1QDPR |
200 |
| HALLMARK_ALLOGRAFT_REJECTION | 3.20e-01 | 2.64 | 0.07 | 1.00e+00 | 1.00e+00 | 1CARTPT |
200 |
| HALLMARK_KRAS_SIGNALING_DN | 3.20e-01 | 2.64 | 0.07 | 1.00e+00 | 1.00e+00 | 1PNMT |
200 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
200 |
| HALLMARK_HYPOXIA | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
200 |
| HALLMARK_CHOLESTEROL_HOMEOSTASIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
74 |
| HALLMARK_MITOTIC_SPINDLE | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
199 |
| HALLMARK_WNT_BETA_CATENIN_SIGNALING | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
42 |
| HALLMARK_TGF_BETA_SIGNALING | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
54 |
| HALLMARK_IL6_JAK_STAT3_SIGNALING | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
87 |
Dowload full table
KEGG Pathways:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| KEGG_TYROSINE_METABOLISM | 7.41e-05 | 42.10 | 8.05 | 1.38e-02 | 1.38e-02 | 3TH, DBH, PNMT |
42 |
| KEGG_FOLATE_BIOSYNTHESIS | 2.11e-02 | 52.51 | 1.19 | 1.00e+00 | 1.00e+00 | 1QDPR |
11 |
| KEGG_PARKINSONS_DISEASE | 2.61e-02 | 8.38 | 0.98 | 1.00e+00 | 1.00e+00 | 2TH, SLC18A1 |
130 |
| KEGG_RENIN_ANGIOTENSIN_SYSTEM | 3.24e-02 | 32.79 | 0.77 | 1.00e+00 | 1.00e+00 | 1ACE |
17 |
| KEGG_NOD_LIKE_RECEPTOR_SIGNALING_PATHWAY | 1.13e-01 | 8.62 | 0.21 | 1.00e+00 | 1.00e+00 | 1NLRP1 |
62 |
| KEGG_HYPERTROPHIC_CARDIOMYOPATHY_HCM | 1.48e-01 | 6.41 | 0.16 | 1.00e+00 | 1.00e+00 | 1ACE |
83 |
| KEGG_VASCULAR_SMOOTH_MUSCLE_CONTRACTION | 1.99e-01 | 4.61 | 0.11 | 1.00e+00 | 1.00e+00 | 1RAMP1 |
115 |
| KEGG_AXON_GUIDANCE | 2.21e-01 | 4.11 | 0.10 | 1.00e+00 | 1.00e+00 | 1UNC5A |
129 |
| KEGG_CELL_ADHESION_MOLECULES_CAMS | 2.27e-01 | 3.98 | 0.10 | 1.00e+00 | 1.00e+00 | 1CNTN1 |
133 |
| KEGG_NEUROACTIVE_LIGAND_RECEPTOR_INTERACTION | 4.08e-01 | 1.94 | 0.05 | 1.00e+00 | 1.00e+00 | 1CHRNA3 |
272 |
| KEGG_N_GLYCAN_BIOSYNTHESIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
46 |
| KEGG_OTHER_GLYCAN_DEGRADATION | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
16 |
| KEGG_O_GLYCAN_BIOSYNTHESIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
30 |
| KEGG_GLYCOSAMINOGLYCAN_DEGRADATION | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
21 |
| KEGG_GLYCOSAMINOGLYCAN_BIOSYNTHESIS_KERATAN_SULFATE | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
15 |
| KEGG_GLYCEROLIPID_METABOLISM | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
49 |
| KEGG_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI_ANCHOR_BIOSYNTHESIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
25 |
| KEGG_GLYCEROPHOSPHOLIPID_METABOLISM | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
77 |
| KEGG_ETHER_LIPID_METABOLISM | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
33 |
| KEGG_ARACHIDONIC_ACID_METABOLISM | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
58 |
Dowload full table
CHR Positional Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| chr20p12 | 1.72e-02 | 10.52 | 1.22 | 1.00e+00 | 1.00e+00 | 2CHGB, PCSK2 |
104 |
| chr6q23 | 1.79e-02 | 10.32 | 1.20 | 1.00e+00 | 1.00e+00 | 2ARFGEF3, SLC35D3 |
106 |
| chr17q23 | 1.98e-02 | 9.76 | 1.13 | 1.00e+00 | 1.00e+00 | 2CYB561, ACE |
112 |
| chr15q22 | 2.39e-02 | 8.80 | 1.02 | 1.00e+00 | 1.00e+00 | 2C2CD4A, HACD3 |
124 |
| chr20p11 | 3.19e-02 | 7.51 | 0.88 | 1.00e+00 | 1.00e+00 | 2INSM1, CFAP61 |
145 |
| chr14q32 | 8.49e-02 | 3.03 | 0.60 | 1.00e+00 | 1.00e+00 | 3CHGA, MEG3, DLK1 |
546 |
| chr1q23 | 6.53e-02 | 4.99 | 0.58 | 1.00e+00 | 1.00e+00 | 2RGS4, RGS5 |
217 |
| chr3p21 | 1.15e-01 | 3.55 | 0.42 | 1.00e+00 | 1.00e+00 | 2NT5DC2, P4HTM |
304 |
| chrXp11 | 1.57e-01 | 2.92 | 0.34 | 1.00e+00 | 1.00e+00 | 2PCSK1N, DGKK |
370 |
| chr12q12 | 1.06e-01 | 9.22 | 0.23 | 1.00e+00 | 1.00e+00 | 1CNTN1 |
58 |
| chr4q22 | 1.27e-01 | 7.62 | 0.19 | 1.00e+00 | 1.00e+00 | 1CCSER1 |
70 |
| chr8q11 | 1.27e-01 | 7.62 | 0.19 | 1.00e+00 | 1.00e+00 | 1ST18 |
70 |
| chr18q11 | 1.43e-01 | 6.65 | 0.16 | 1.00e+00 | 1.00e+00 | 1ANKRD29 |
80 |
| chr2q36 | 1.47e-01 | 6.49 | 0.16 | 1.00e+00 | 1.00e+00 | 1SCG2 |
82 |
| chr15q13 | 1.55e-01 | 6.11 | 0.15 | 1.00e+00 | 1.00e+00 | 1SCG5 |
87 |
| chr8q12 | 1.57e-01 | 6.04 | 0.15 | 1.00e+00 | 1.00e+00 | 1PENK |
88 |
| chrXq23 | 1.58e-01 | 5.97 | 0.15 | 1.00e+00 | 1.00e+00 | 1SERTM2 |
89 |
| chr18q12 | 1.70e-01 | 5.53 | 0.14 | 1.00e+00 | 1.00e+00 | 1SYT4 |
96 |
| chr3q27 | 1.99e-01 | 4.61 | 0.11 | 1.00e+00 | 1.00e+00 | 1VWA5B2 |
115 |
| chr6q22 | 2.06e-01 | 4.46 | 0.11 | 1.00e+00 | 1.00e+00 | 1FAM162B |
119 |
Dowload full table
Transcription Factor Targets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| NRSF_01 | 3.73e-05 | 24.08 | 6.17 | 1.41e-02 | 4.22e-02 | 4CHGA, PCSK2, VGF, SYT4 |
97 |
| MIER1_TARGET_GENES | 5.22e-07 | 13.37 | 5.38 | 5.91e-04 | 5.91e-04 | 8CHGB, SCG2, CARTPT, VWA5B2, MIR7-3HG, SEZ6L2, VGF, CNIH2 |
375 |
| ATF_B | 3.29e-05 | 15.39 | 4.71 | 1.41e-02 | 3.73e-02 | 5CHGB, SCG2, TH, SEZ6L2, VGF |
191 |
| TGACGTCA_ATF3_Q6 | 8.72e-05 | 12.44 | 3.82 | 1.71e-02 | 9.88e-02 | 5CHGB, SCG2, TH, SEZ6L2, VGF |
235 |
| ATF1_Q6 | 9.07e-05 | 12.34 | 3.79 | 1.71e-02 | 1.03e-01 | 5CHGB, SCG2, TH, SEZ6L2, VGF |
237 |
| CAGNWMCNNNGAC_UNKNOWN | 6.62e-04 | 19.34 | 3.78 | 7.50e-02 | 7.50e-01 | 3CHGA, CHGB, SYT4 |
88 |
| CREBP1_Q2 | 1.39e-04 | 11.23 | 3.45 | 2.02e-02 | 1.58e-01 | 5CHGB, SCG2, TH, SEZ6L2, VGF |
260 |
| CREBP1CJUN_01 | 1.50e-04 | 11.05 | 3.40 | 2.02e-02 | 1.69e-01 | 5CHGB, SCG2, TH, SEZ6L2, VGF |
264 |
| CREB_01 | 1.60e-04 | 10.88 | 3.34 | 2.02e-02 | 1.82e-01 | 5CHGB, SCG2, TH, SEZ6L2, VGF |
268 |
| BAHD1_TARGET_GENES | 2.60e-03 | 28.99 | 3.29 | 1.47e-01 | 1.00e+00 | 2PENK, MIR7-3HG |
39 |
| TGAYRTCA_ATF3_Q6 | 7.52e-05 | 7.74 | 2.92 | 1.71e-02 | 8.52e-02 | 7CHGB, SCG2, TH, CARTPT, PCSK2, SEZ6L2, VGF |
549 |
| CREB_Q4_01 | 8.01e-04 | 10.51 | 2.72 | 8.26e-02 | 9.08e-01 | 4CHGB, SCG2, TH, SEZ6L2 |
217 |
| GGGNNTTTCC_NFKB_Q6_01 | 2.21e-03 | 12.55 | 2.47 | 1.32e-01 | 1.00e+00 | 3C1QL1, PCSK2, CNIH2 |
134 |
| ATF3_Q6 | 1.41e-03 | 8.99 | 2.33 | 1.15e-01 | 1.00e+00 | 4CHGB, SCG2, INSM1, SEZ6L2 |
253 |
| CREB_02 | 1.55e-03 | 8.75 | 2.27 | 1.15e-01 | 1.00e+00 | 4CHGB, SCG2, PCSK2, SEZ6L2 |
260 |
| ATF_01 | 1.71e-03 | 8.52 | 2.21 | 1.15e-01 | 1.00e+00 | 4CHGB, SCG2, TH, SEZ6L2 |
267 |
| CREB_Q2 | 1.76e-03 | 8.45 | 2.19 | 1.15e-01 | 1.00e+00 | 4CHGB, SCG2, PENK, SEZ6L2 |
269 |
| EGR1_01 | 1.76e-03 | 8.45 | 2.19 | 1.15e-01 | 1.00e+00 | 4PCSK2, SEZ6L2, VGF, ACE |
269 |
| CREB_Q4 | 1.83e-03 | 8.36 | 2.17 | 1.15e-01 | 1.00e+00 | 4CHGB, SCG2, PENK, SEZ6L2 |
272 |
| RBMX_TARGET_GENES | 1.35e-02 | 87.42 | 1.87 | 5.39e-01 | 1.00e+00 | 1MIR7-3HG |
7 |
Dowload full table
GO Biological Processes:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GOBP_NOREPINEPHRINE_BIOSYNTHETIC_PROCESS | 2.38e-07 | 406.69 | 58.21 | 4.45e-04 | 1.78e-03 | 3INSM1, TH, DBH |
7 |
| GOBP_CATECHOL_CONTAINING_COMPOUND_BIOSYNTHETIC_PROCESS | 5.88e-08 | 139.68 | 32.68 | 1.74e-04 | 4.40e-04 | 4INSM1, TH, DBH, PNMT |
20 |
| GOBP_NOREPINEPHRINE_METABOLIC_PROCESS | 1.93e-06 | 163.67 | 28.07 | 1.80e-03 | 1.44e-02 | 3INSM1, TH, DBH |
13 |
| GOBP_SIGNALING_RECEPTOR_LIGAND_PRECURSOR_PROCESSING | 4.87e-07 | 77.25 | 18.93 | 7.28e-04 | 3.64e-03 | 4PCSK1N, SCG5, PCSK2, ACE |
33 |
| GOBP_AMINE_TRANSPORT | 5.80e-10 | 48.13 | 17.78 | 4.34e-06 | 4.34e-06 | 7CHGA, TH, CARTPT, SLC18A1, CHRNA3, RGS4, SYT4 |
94 |
| GOBP_NEUROTRANSMITTER_LOADING_INTO_SYNAPTIC_VESICLE | 1.02e-04 | 177.85 | 17.16 | 2.65e-02 | 7.64e-01 | 2TH, SLC18A1 |
8 |
| GOBP_AMINE_BIOSYNTHETIC_PROCESS | 9.69e-07 | 63.88 | 15.85 | 1.21e-03 | 7.25e-03 | 4INSM1, TH, DBH, PNMT |
39 |
| GOBP_PHENOL_CONTAINING_COMPOUND_BIOSYNTHETIC_PROCESS | 1.44e-06 | 57.35 | 14.31 | 1.54e-03 | 1.08e-02 | 4INSM1, TH, DBH, PNMT |
43 |
| GOBP_NEGATIVE_REGULATION_OF_AMINE_TRANSPORT | 1.72e-05 | 71.33 | 13.27 | 7.32e-03 | 1.29e-01 | 3CHGA, RGS4, SYT4 |
26 |
| GOBP_CATECHOL_CONTAINING_COMPOUND_METABOLIC_PROCESS | 3.12e-06 | 46.57 | 11.73 | 2.60e-03 | 2.34e-02 | 4INSM1, TH, DBH, PNMT |
52 |
| GOBP_RESPONSE_TO_AMPHETAMINE | 2.67e-05 | 60.79 | 11.42 | 1.05e-02 | 2.00e-01 | 3TH, DBH, RGS4 |
30 |
| GOBP_NEGATIVE_REGULATION_OF_CATECHOLAMINE_SECRETION | 2.83e-04 | 97.16 | 10.22 | 5.33e-02 | 1.00e+00 | 2CHGA, SYT4 |
13 |
| GOBP_GANGLION_DEVELOPMENT | 4.33e-04 | 76.37 | 8.22 | 7.90e-02 | 1.00e+00 | 2INSM1, RGS4 |
16 |
| GOBP_RESPONSE_TO_AMINE | 7.41e-05 | 42.10 | 8.05 | 2.13e-02 | 5.54e-01 | 3TH, DBH, RGS4 |
42 |
| GOBP_RESPONSE_TO_NICOTINE | 7.95e-05 | 41.01 | 7.86 | 2.20e-02 | 5.95e-01 | 3PENK, TH, CHRNA3 |
43 |
| GOBP_MONOAMINE_TRANSPORT | 1.66e-05 | 29.85 | 7.61 | 7.32e-03 | 1.24e-01 | 4CHGA, CARTPT, SLC18A1, SYT4 |
79 |
| GOBP_NEUROTRANSMITTER_BIOSYNTHETIC_PROCESS | 5.51e-04 | 67.02 | 7.27 | 9.81e-02 | 1.00e+00 | 2TH, DBH |
18 |
| GOBP_LEARNING | 5.17e-06 | 22.87 | 6.97 | 3.87e-03 | 3.87e-02 | 5C1QL1, TH, DBH, ARC, SLC24A2 |
130 |
| GOBP_CATECHOLAMINE_SECRETION | 1.49e-04 | 32.83 | 6.33 | 3.59e-02 | 1.00e+00 | 3CHGA, CARTPT, SYT4 |
53 |
| GOBP_CELLULAR_BIOGENIC_AMINE_METABOLIC_PROCESS | 4.71e-05 | 22.60 | 5.80 | 1.63e-02 | 3.52e-01 | 4INSM1, TH, DBH, PNMT |
103 |
Dowload full table
Immunological Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GSE43956_WT_VS_SGK1_KO_TH17_DIFFERENTIATED_CD4_TCELL_UP | 4.09e-05 | 14.68 | 4.50 | 1.99e-01 | 1.99e-01 | 5CHGB, PENK, CYB561, QDPR, MEG3 |
200 |
| GSE9006_1MONTH_VS_4MONTH_POST_TYPE_1_DIABETES_DX_PBMC_DN | 5.39e-04 | 11.73 | 3.04 | 2.62e-01 | 1.00e+00 | 4RAMP1, CHRNA3, SLC24A2, PCSK2 |
195 |
| GSE20727_ROS_INH_VS_ROS_INH_AND_DNFB_ALLERGEN_TREATED_DC_DN | 5.49e-04 | 11.66 | 3.02 | 2.62e-01 | 1.00e+00 | 4CARTPT, CHRNA3, CNIH2, CFAP61 |
196 |
| GSE41176_UNSTIM_VS_ANTI_IGM_STIM_BCELL_24H_UP | 5.71e-04 | 11.54 | 2.99 | 2.62e-01 | 1.00e+00 | 4SCG2, RAMP1, DBH, CNTN1 |
198 |
| GSE12845_IGD_POS_VS_NEG_BLOOD_BCELL_UP | 5.81e-04 | 11.49 | 2.97 | 2.62e-01 | 1.00e+00 | 4RAMP1, PENK, DBH, NLRP1 |
199 |
| GSE45365_NK_CELL_VS_CD8A_DC_DN | 5.81e-04 | 11.49 | 2.97 | 2.62e-01 | 1.00e+00 | 4TH, CNTN1, DGKK, SEZ6L2 |
199 |
| GSE13229_MATURE_VS_INTMATURE_NKCELL_UP | 5.92e-04 | 11.43 | 2.96 | 2.62e-01 | 1.00e+00 | 4RAMP1, PENK, CCSER1, ACE |
200 |
| GSE20366_EX_VIVO_VS_HOMEOSTATIC_CONVERSION_NAIVE_CD4_TCELL_DN | 5.92e-04 | 11.43 | 2.96 | 2.62e-01 | 1.00e+00 | 4PNMT, SLC35D3, DLK1, ZDBF2 |
200 |
| GSE22886_DC_VS_MONOCYTE_UP | 5.92e-04 | 11.43 | 2.96 | 2.62e-01 | 1.00e+00 | 4RAMP1, NT5DC2, ACE, HACD3 |
200 |
| GSE4142_NAIVE_VS_MEMORY_BCELL_DN | 5.92e-04 | 11.43 | 2.96 | 2.62e-01 | 1.00e+00 | 4PNMT, SLC24A2, FAM162B, HACD3 |
200 |
| GSE23502_BM_VS_COLON_TUMOR_HDC_KO_MYELOID_DERIVED_SUPPRESSOR_CELL_UP | 5.92e-04 | 11.43 | 2.96 | 2.62e-01 | 1.00e+00 | 4SLC18A1, CHRNA3, SLC24A2, SEZ6L2 |
200 |
| GSE12963_UNINF_VS_ENV_AND_NEF_DEFICIENT_HIV1_INF_CD4_TCELL_UP | 2.55e-03 | 11.91 | 2.34 | 7.45e-01 | 1.00e+00 | 3CHGB, TH, PNMT |
141 |
| GSE40274_CTRL_VS_FOXP3_AND_SATB1_TRANSDUCED_ACTIVATED_CD4_TCELL_DN | 3.83e-03 | 10.27 | 2.02 | 7.45e-01 | 1.00e+00 | 3CHGA, TH, FAM162B |
163 |
| GSE40274_CTRL_VS_LEF1_TRANSDUCED_ACTIVATED_CD4_TCELL_DN | 4.31e-03 | 9.84 | 1.94 | 7.45e-01 | 1.00e+00 | 3PENK, ST18, CNIH2 |
170 |
| GSE15215_CD2_POS_VS_NEG_PDC_UP | 6.37e-03 | 8.52 | 1.68 | 7.45e-01 | 1.00e+00 | 3NT5DC2, NLRP1, ACE |
196 |
| GSE22886_NEUTROPHIL_VS_MONOCYTE_UP | 6.46e-03 | 8.47 | 1.67 | 7.45e-01 | 1.00e+00 | 3CHRNA3, RGS4, VGF |
197 |
| GSE9006_TYPE_1_DIABETES_AT_DX_VS_1MONTH_POST_DX_PBMC_DN | 6.46e-03 | 8.47 | 1.67 | 7.45e-01 | 1.00e+00 | 3SLC18A1, PCSK2, RGS5 |
197 |
| GSE19401_PAM2CSK4_VS_RETINOIC_ACID_STIM_FOLLICULAR_DC_DN | 6.46e-03 | 8.47 | 1.67 | 7.45e-01 | 1.00e+00 | 3SCG2, DBH, CNTN1 |
197 |
| GSE20500_RETINOIC_ACID_VS_RARA_ANTAGONIST_TREATED_CD4_TCELL_DN | 6.55e-03 | 8.43 | 1.66 | 7.45e-01 | 1.00e+00 | 3CHGA, SCG2, SCG5 |
198 |
| GSE11961_PLASMA_CELL_DAY7_VS_MEMORY_BCELL_DAY40_DN | 6.55e-03 | 8.43 | 1.66 | 7.45e-01 | 1.00e+00 | 3SLC18A1, ARFGEF3, CNIH2 |
198 |
Top Ranked Transcription Factors for this Gene Expression Program:
| Gene Symbol | TF Rank | DNA Binding Domain | Motif Status | IUPAC PWM | GTEx | DepMap | Decartes |
|---|---|---|---|---|---|---|---|
| INSM1 | 6 | Yes | Known motif | Monomer or homomultimer | In vivo/Misc source | None | None |
| ST18 | 38 | Yes | Inferred motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| ADGRA1 | 60 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| ZNF331 | 81 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | ChIP-seq motif is consistent with recognition code (RCADE) | None |
| INS | 87 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| NPAS4 | 88 | Yes | Inferred motif | Obligate heteromer | High-throughput in vitro | None | Likely dimerizes with ARNT2 (PMID:24263188). |
| BEX1 | 100 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Likely a cofactor- (PMID: 16314316) shows results for related BEX2 |
| HAND2 | 113 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| NR4A1 | 156 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| GPR155 | 162 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| PHOX2A | 166 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| NKX6-2 | 180 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | Has a putative AT-hook | None |
| TRIM29 | 183 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Previous evidence for TF activity (PMID: 8188213) is based on sequence analysis and does not provide DNA-binding evidence. Based on EntrezGene information, its likely a transcriptional co-factor |
| KLF13 | 185 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| TLX2 | 187 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| BTG2 | 190 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Transcriptional co-factor (PMID: 10617598) |
| VEGFA | 200 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Extracellular signaling molecule. Included only because it regulates CREB. |
| JUND | 203 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| TBX20 | 227 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| HMGN3 | 237 | Yes | Likely to be sequence specific TF | Low specificity DNA-binding protein | No motif | None | None |
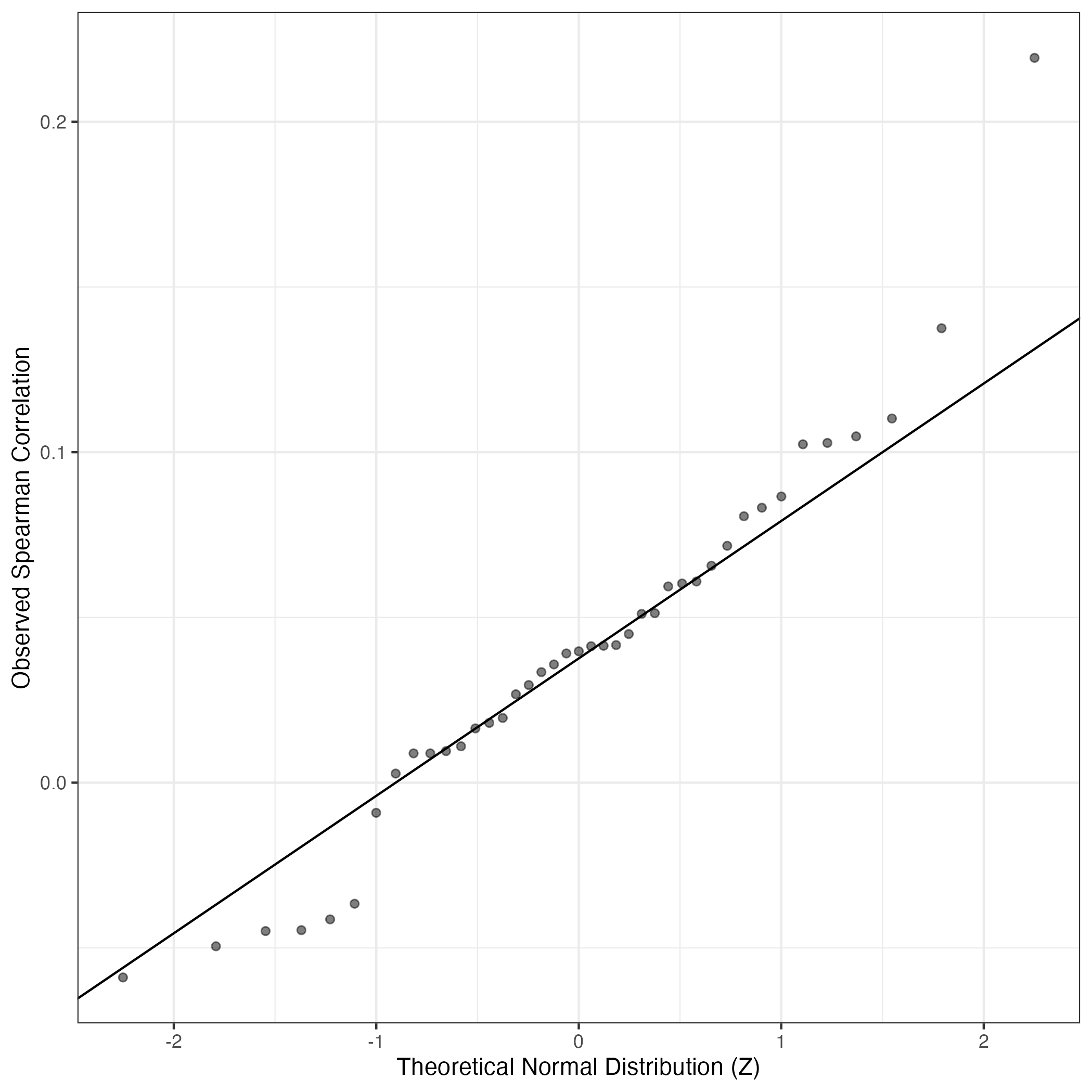
QQ Plot showing correlations with other GEPs in this dataset, calculated by Spearman correlation:
Interactive QQ-plot of gene loadings:
A similar QQ-plot as above, but only for instances where the H value is e.g. > 25, i.e. we are confident that the expression program is active above noise. Agreemenet between these binary vectors is tested using the Jaccard Index, with the P-values calculated by an exact test:
Interactive QQ-plot:
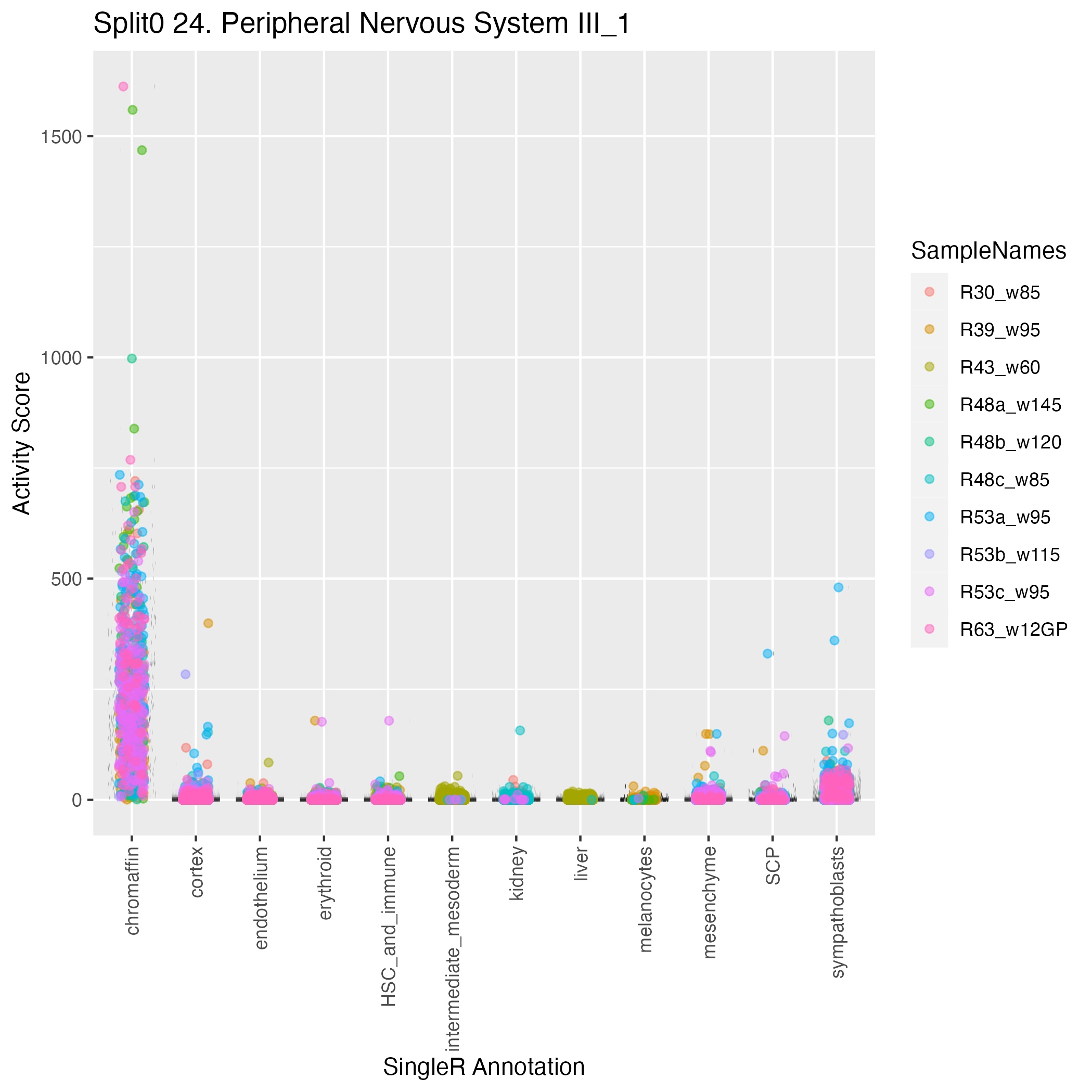
Singler cell type annotations for the top 50 cells on this program.
| Cell ID | Singler label | Singler Delta | Activity Score | Top Singler Raw Scores |
|---|---|---|---|---|
| R63_w12GP_GATGAGGCAACTTCTT-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 1612.37 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.44, Neuroepithelial_cell:ESC-derived: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.37, iPS_cells:PDB_2lox-22: 0.37, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.37, iPS_cells:PDB_1lox-17Puro-10: 0.37, Embryonic_stem_cells: 0.37, iPS_cells:PDB_1lox-21Puro-26: 0.37, iPS_cells:PDB_2lox-17: 0.37 |
| R48a_w14.5_TGGCGTGAGGTTATAG-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 1559.46 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.47, Neuroepithelial_cell:ESC-derived: 0.42, Astrocyte:Embryonic_stem_cell-derived: 0.41, Neurons:Schwann_cell: 0.4, iPS_cells:PDB_1lox-17Puro-10: 0.4, iPS_cells:PDB_1lox-21Puro-20: 0.4, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.4, iPS_cells:skin_fibroblast-derived: 0.4, iPS_cells:PDB_2lox-5: 0.4, Embryonic_stem_cells: 0.39 |
| R48a_w14.5_ATCTCTATCCTTCACG-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 1468.39 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.43, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.37, iPS_cells:PDB_1lox-21Puro-20: 0.36, iPS_cells:PDB_1lox-17Puro-10: 0.36, iPS_cells:PDB_1lox-21Puro-26: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.35, iPS_cells:PDB_2lox-5: 0.35, iPS_cells:PDB_2lox-22: 0.35, iPS_cells:PDB_2lox-17: 0.35 |
| R48b_w12_GGTGTCGAGGTGCTTT-1 | Neurons:adrenal_medulla_cell_line | 0.20 | 997.47 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.5, Neuroepithelial_cell:ESC-derived: 0.43, Astrocyte:Embryonic_stem_cell-derived: 0.42, iPS_cells:PDB_1lox-17Puro-10: 0.41, iPS_cells:PDB_1lox-17Puro-5: 0.41, iPS_cells:PDB_1lox-21Puro-20: 0.41, iPS_cells:PDB_1lox-21Puro-26: 0.41, iPS_cells:PDB_2lox-22: 0.4, Embryonic_stem_cells: 0.4, iPS_cells:PDB_2lox-17: 0.4 |
| R48a_w14.5_ATCGTGAGTTTAGACC-1 | Neurons:adrenal_medulla_cell_line | 0.19 | 838.93 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.46, Neuroepithelial_cell:ESC-derived: 0.41, Astrocyte:Embryonic_stem_cell-derived: 0.39, iPS_cells:PDB_1lox-17Puro-5: 0.39, iPS_cells:PDB_1lox-17Puro-10: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.38, iPS_cells:PDB_1lox-21Puro-26: 0.38, iPS_cells:PDB_2lox-22: 0.38, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.38, iPS_cells:PDB_2lox-5: 0.38 |
| R63_w12GP_GTTCCGTGTTGCTGAT-1 | Neurons:adrenal_medulla_cell_line | 0.13 | 768.49 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.37, Neuroepithelial_cell:ESC-derived: 0.34, Astrocyte:Embryonic_stem_cell-derived: 0.33, iPS_cells:PDB_2lox-5: 0.32, iPS_cells:PDB_2lox-22: 0.32, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.32, iPS_cells:iPS:minicircle-derived: 0.32, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.32, iPS_cells:PDB_2lox-17: 0.32, iPS_cells:PDB_2lox-21: 0.32 |
| R53a_w9.5_CTTCAATTCTGTACAG-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 734.65 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.39, Neuroepithelial_cell:ESC-derived: 0.34, Astrocyte:Embryonic_stem_cell-derived: 0.33, Neurons:ES_cell-derived_neural_precursor: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.32, iPS_cells:PDB_1lox-21Puro-20: 0.32, iPS_cells:PDB_1lox-21Puro-26: 0.32, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.32, iPS_cells:PDB_2lox-22: 0.32 |
| R30_w8.5_CGTAATGTCTGGGCAC-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 720.78 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.42, Neuroepithelial_cell:ESC-derived: 0.37, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.35, iPS_cells:PDB_2lox-22: 0.35, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.35, iPS_cells:PDB_2lox-5: 0.35, iPS_cells:PDB_2lox-17: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.35, iPS_cells:iPS:minicircle-derived: 0.35 |
| R53a_w9.5_TTTACCAGTGAGCCAA-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 712.33 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.45, Neuroepithelial_cell:ESC-derived: 0.43, iPS_cells:PDB_1lox-17Puro-10: 0.41, iPS_cells:PDB_1lox-17Puro-5: 0.41, Neurons:ES_cell-derived_neural_precursor: 0.41, iPS_cells:PDB_1lox-21Puro-26: 0.41, iPS_cells:PDB_1lox-21Puro-20: 0.41, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.41, iPS_cells:PDB_2lox-22: 0.4, Astrocyte:Embryonic_stem_cell-derived: 0.4 |
| R63_w12GP_GAAGGGTGTGGAACAC-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 707.57 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.42, Neuroepithelial_cell:ESC-derived: 0.37, iPS_cells:PDB_1lox-17Puro-5: 0.37, iPS_cells:PDB_1lox-21Puro-20: 0.37, iPS_cells:PDB_1lox-17Puro-10: 0.36, iPS_cells:PDB_1lox-21Puro-26: 0.36, Astrocyte:Embryonic_stem_cell-derived: 0.36, iPS_cells:PDB_2lox-22: 0.36, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.36, iPS_cells:PDB_2lox-17: 0.36 |
| R63_w12GP_CATACCCAGCCATATC-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 707.41 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.31, Neuroepithelial_cell:ESC-derived: 0.28, Astrocyte:Embryonic_stem_cell-derived: 0.26, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.26, Neurons:ES_cell-derived_neural_precursor: 0.26, iPS_cells:PDB_2lox-5: 0.25, Embryonic_stem_cells: 0.25, iPS_cells:PDB_1lox-17Puro-10: 0.25, Neurons:Schwann_cell: 0.25, iPS_cells:PDB_1lox-17Puro-5: 0.25 |
| R53a_w9.5_GTCAAGTCACCCTTAC-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 688.19 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.41, Neuroepithelial_cell:ESC-derived: 0.37, Astrocyte:Embryonic_stem_cell-derived: 0.36, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.34, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_2lox-22: 0.34, Neurons:ES_cell-derived_neural_precursor: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_2lox-21: 0.34 |
| R48a_w14.5_CTTCAATTCGTTCTCG-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 686.34 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.4, Neuroepithelial_cell:ESC-derived: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.35, Neurons:Schwann_cell: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.33, iPS_cells:PDB_1lox-21Puro-20: 0.33, iPS_cells:PDB_1lox-21Puro-26: 0.33, iPS_cells:PDB_2lox-22: 0.33, Embryonic_stem_cells: 0.33 |
| R53a_w9.5_TCAATTCGTATGCTTG-1 | Neurons:adrenal_medulla_cell_line | 0.18 | 684.80 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.47, Neuroepithelial_cell:ESC-derived: 0.44, iPS_cells:PDB_1lox-21Puro-20: 0.42, iPS_cells:PDB_1lox-17Puro-5: 0.42, iPS_cells:PDB_1lox-17Puro-10: 0.42, iPS_cells:PDB_1lox-21Puro-26: 0.42, Astrocyte:Embryonic_stem_cell-derived: 0.41, iPS_cells:PDB_2lox-22: 0.41, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.41, iPS_cells:PDB_2lox-5: 0.41 |
| R48a_w14.5_CTACTATTCTTGCGCT-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 681.86 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.42, Neuroepithelial_cell:ESC-derived: 0.36, Astrocyte:Embryonic_stem_cell-derived: 0.34, MSC: 0.34, Neurons:Schwann_cell: 0.34, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.34, iPS_cells:iPS:minicircle-derived: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:skin_fibroblast-derived: 0.34 |
| R48c_w8.5_ATCGCCTTCCTCGCAT-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 674.93 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.42, Neurons:adrenal_medulla_cell_line: 0.42, Astrocyte:Embryonic_stem_cell-derived: 0.4, Neurons:ES_cell-derived_neural_precursor: 0.4, Neurons:Schwann_cell: 0.39, Embryonic_stem_cells: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.38, iPS_cells:PDB_1lox-17Puro-10: 0.38, iPS_cells:PDB_1lox-21Puro-26: 0.37, iPS_cells:PDB_1lox-17Puro-5: 0.37 |
| R48a_w14.5_CCGAACGTCTAATTCC-1 | Neurons:adrenal_medulla_cell_line | 0.18 | 672.79 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.45, Neuroepithelial_cell:ESC-derived: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-17Puro-10: 0.37, iPS_cells:PDB_1lox-21Puro-26: 0.37, iPS_cells:PDB_1lox-21Puro-20: 0.37, iPS_cells:PDB_1lox-17Puro-5: 0.37, Neurons:Schwann_cell: 0.36, iPS_cells:PDB_2lox-22: 0.36, iPS_cells:PDB_2lox-5: 0.36 |
| R53a_w9.5_CCGGTGATCTCTCGCA-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 671.81 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.4, Neuroepithelial_cell:ESC-derived: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.33, iPS_cells:PDB_1lox-21Puro-20: 0.33, Neurons:ES_cell-derived_neural_precursor: 0.33, iPS_cells:PDB_1lox-21Puro-26: 0.33, iPS_cells:PDB_2lox-22: 0.33, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.32 |
| R48a_w14.5_GATGCTAGTGGTACAG-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 662.70 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.38, Neuroepithelial_cell:ESC-derived: 0.34, Astrocyte:Embryonic_stem_cell-derived: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.32, iPS_cells:skin_fibroblast-derived: 0.32, iPS_cells:PDB_1lox-21Puro-20: 0.32, iPS_cells:PDB_1lox-17Puro-5: 0.32, iPS_cells:PDB_1lox-21Puro-26: 0.32, Embryonic_stem_cells: 0.32, iPS_cells:iPS:minicircle-derived: 0.32 |
| R30_w8.5_TCGCTCATCAGACATC-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 655.91 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.34, Neuroepithelial_cell:ESC-derived: 0.29, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.29, Astrocyte:Embryonic_stem_cell-derived: 0.29, iPS_cells:PDB_1lox-21Puro-20: 0.28, iPS_cells:PDB_1lox-17Puro-5: 0.28, iPS_cells:PDB_2lox-5: 0.28, iPS_cells:PDB_1lox-17Puro-10: 0.28, iPS_cells:PDB_1lox-21Puro-26: 0.28, iPS_cells:PDB_2lox-22: 0.28 |
| R48a_w14.5_GTGCAGCGTTTCGTGA-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 652.23 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.4, Neuroepithelial_cell:ESC-derived: 0.36, Astrocyte:Embryonic_stem_cell-derived: 0.34, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_2lox-22: 0.34, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.34, iPS_cells:PDB_2lox-5: 0.33 |
| R53c_w9.5_TGTTCTAAGTAGCCAG-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 650.39 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.45, Neuroepithelial_cell:ESC-derived: 0.42, iPS_cells:PDB_1lox-17Puro-5: 0.4, iPS_cells:PDB_1lox-17Puro-10: 0.4, iPS_cells:PDB_1lox-21Puro-20: 0.4, iPS_cells:PDB_1lox-21Puro-26: 0.4, Astrocyte:Embryonic_stem_cell-derived: 0.4, iPS_cells:PDB_2lox-22: 0.4, Neurons:ES_cell-derived_neural_precursor: 0.4, iPS_cells:PDB_2lox-21: 0.39 |
| R48a_w14.5_TTCTTCCTCATCTATC-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 633.93 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.44, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-17Puro-5: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.35, iPS_cells:PDB_1lox-21Puro-26: 0.35, iPS_cells:PDB_2lox-22: 0.35, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.35, iPS_cells:PDB_2lox-5: 0.35 |
| R53a_w9.5_TTTGGTTGTGCCGTTG-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 626.66 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.42, Neuroepithelial_cell:ESC-derived: 0.36, Astrocyte:Embryonic_stem_cell-derived: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.34, iPS_cells:PDB_1lox-21Puro-20: 0.34, Neurons:ES_cell-derived_neural_precursor: 0.34, Embryonic_stem_cells: 0.34 |
| R63_w12GP_CACGGGTGTTTACTGG-1 | Endothelial_cells:blood_vessel | 0.14 | 619.92 | Raw ScoresEndothelial_cells:lymphatic: 0.39, Endothelial_cells:blood_vessel: 0.39, Endothelial_cells:lymphatic:KSHV: 0.39, Endothelial_cells:lymphatic:TNFa_48h: 0.38, Endothelial_cells:HUVEC: 0.38, Endothelial_cells:HUVEC:FPV-infected: 0.38, Endothelial_cells:HUVEC:B._anthracis_LT: 0.37, Endothelial_cells:HUVEC:Serum_Amyloid_A: 0.37, Endothelial_cells:HUVEC:VEGF: 0.37, Endothelial_cells:HUVEC:H5N1-infected: 0.37 |
| R48a_w14.5_ATGTCCCTCTCACCCA-1 | Neurons:adrenal_medulla_cell_line | 0.19 | 611.18 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.44, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.37, iPS_cells:PDB_1lox-17Puro-10: 0.36, iPS_cells:PDB_1lox-21Puro-26: 0.36, iPS_cells:PDB_1lox-21Puro-20: 0.36, iPS_cells:PDB_1lox-17Puro-5: 0.36, iPS_cells:PDB_2lox-22: 0.35, iPS_cells:PDB_2lox-21: 0.35, Neurons:ES_cell-derived_neural_precursor: 0.35 |
| R53a_w9.5_GTACAACCAGCGTTGC-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 605.55 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.4, Neuroepithelial_cell:ESC-derived: 0.35, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.34, Neurons:ES_cell-derived_neural_precursor: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.34, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_2lox-22: 0.34, iPS_cells:PDB_2lox-21: 0.34 |
| R39_w9.5_CAGCCAGTCATCCTGC-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 603.89 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.37, Neuroepithelial_cell:ESC-derived: 0.33, Astrocyte:Embryonic_stem_cell-derived: 0.31, Neurons:Schwann_cell: 0.31, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.31, MSC: 0.31, Embryonic_stem_cells: 0.31, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.31, Tissue_stem_cells:CD326-CD56+: 0.31, iPS_cells:PDB_1lox-17Puro-10: 0.31 |
| R30_w8.5_TTGTGGAGTCTGTAAC-1 | Neurons:adrenal_medulla_cell_line | 0.18 | 602.19 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.44, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.37, iPS_cells:PDB_1lox-17Puro-10: 0.36, iPS_cells:PDB_1lox-17Puro-5: 0.36, iPS_cells:PDB_1lox-21Puro-26: 0.36, iPS_cells:PDB_1lox-21Puro-20: 0.36, iPS_cells:PDB_2lox-22: 0.36, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.36, Embryonic_stem_cells: 0.36 |
| R48a_w14.5_TGAGCATGTCAGGCAA-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 594.47 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.4, Neuroepithelial_cell:ESC-derived: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.33, Neurons:Schwann_cell: 0.32, iPS_cells:PDB_1lox-21Puro-20: 0.32, Embryonic_stem_cells: 0.32, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.32, iPS_cells:skin_fibroblast-derived: 0.32, iPS_cells:PDB_2lox-5: 0.32, iPS_cells:PDB_1lox-21Puro-26: 0.31 |
| R53a_w9.5_TGCCGAGTCCAGGACC-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 590.37 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.4, Neuroepithelial_cell:ESC-derived: 0.34, Astrocyte:Embryonic_stem_cell-derived: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.32, iPS_cells:PDB_1lox-17Puro-10: 0.32, Neurons:ES_cell-derived_neural_precursor: 0.32, iPS_cells:PDB_1lox-21Puro-20: 0.32, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.32, iPS_cells:PDB_1lox-21Puro-26: 0.32, iPS_cells:PDB_2lox-22: 0.31 |
| R63_w12GP_ACGTAGTTCACTTTGT-1 | Neurons:adrenal_medulla_cell_line | 0.13 | 586.85 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.38, Neuroepithelial_cell:ESC-derived: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.32, iPS_cells:PDB_1lox-17Puro-10: 0.32, iPS_cells:iPS:minicircle-derived: 0.32, iPS_cells:PDB_1lox-21Puro-20: 0.32, iPS_cells:PDB_2lox-22: 0.32, iPS_cells:PDB_1lox-21Puro-26: 0.32, iPS_cells:PDB_2lox-21: 0.32, iPS_cells:PDB_2lox-17: 0.32 |
| R53a_w9.5_TCAAGTGCACGCGCTA-1 | Neurons:adrenal_medulla_cell_line | 0.21 | 578.76 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.52, Neuroepithelial_cell:ESC-derived: 0.49, iPS_cells:PDB_2lox-22: 0.48, iPS_cells:PDB_1lox-17Puro-10: 0.48, iPS_cells:PDB_1lox-21Puro-20: 0.48, iPS_cells:PDB_1lox-17Puro-5: 0.48, iPS_cells:PDB_1lox-21Puro-26: 0.48, iPS_cells:PDB_2lox-17: 0.47, iPS_cells:PDB_2lox-21: 0.47, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.47 |
| R48a_w14.5_CTTCCGAAGCCTTTGA-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 574.40 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.37, Neuroepithelial_cell:ESC-derived: 0.33, Astrocyte:Embryonic_stem_cell-derived: 0.33, Embryonic_stem_cells: 0.3, iPS_cells:PDB_1lox-21Puro-20: 0.3, iPS_cells:PDB_1lox-21Puro-26: 0.3, iPS_cells:PDB_1lox-17Puro-10: 0.3, iPS_cells:PDB_1lox-17Puro-5: 0.3, iPS_cells:PDB_2lox-22: 0.3, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.3 |
| R48b_w12_TCTGCCAGTGTCTAAC-1 | Neurons:adrenal_medulla_cell_line | 0.18 | 571.46 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.43, Neuroepithelial_cell:ESC-derived: 0.37, Astrocyte:Embryonic_stem_cell-derived: 0.36, iPS_cells:PDB_1lox-17Puro-10: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.35, iPS_cells:PDB_1lox-21Puro-26: 0.35, iPS_cells:PDB_2lox-22: 0.34, iPS_cells:PDB_2lox-17: 0.34, Embryonic_stem_cells: 0.34 |
| R63_w12GP_CGTAGTACAGAAGCGT-1 | Neurons:adrenal_medulla_cell_line | 0.12 | 567.04 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Neuroepithelial_cell:ESC-derived: 0.28, Astrocyte:Embryonic_stem_cell-derived: 0.28, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.27, iPS_cells:iPS:minicircle-derived: 0.27, iPS_cells:PDB_2lox-5: 0.27, iPS_cells:PDB_2lox-22: 0.27, iPS_cells:PDB_2lox-17: 0.27, iPS_cells:skin_fibroblast-derived: 0.27, iPS_cells:adipose_stem_cell-derived:lentiviral: 0.27 |
| R53a_w9.5_TCTCTGGCAACTCGAT-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 566.57 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.43, Neuroepithelial_cell:ESC-derived: 0.38, iPS_cells:PDB_1lox-17Puro-10: 0.36, iPS_cells:PDB_1lox-17Puro-5: 0.36, iPS_cells:PDB_1lox-21Puro-20: 0.36, iPS_cells:PDB_1lox-21Puro-26: 0.36, iPS_cells:PDB_2lox-22: 0.36, iPS_cells:PDB_2lox-17: 0.36, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.36, iPS_cells:PDB_2lox-21: 0.35 |
| R63_w12GP_CCTCTAGTCGTGCACG-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 564.48 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.39, Neuroepithelial_cell:ESC-derived: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.34, iPS_cells:PDB_1lox-21Puro-20: 0.34, Astrocyte:Embryonic_stem_cell-derived: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_2lox-22: 0.33, iPS_cells:PDB_2lox-5: 0.33 |
| R53a_w9.5_CATTCCGAGTCAGCGA-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 563.36 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.43, Neuroepithelial_cell:ESC-derived: 0.39, iPS_cells:PDB_2lox-22: 0.38, iPS_cells:PDB_2lox-5: 0.38, iPS_cells:PDB_1lox-17Puro-5: 0.38, iPS_cells:PDB_1lox-17Puro-10: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.38, iPS_cells:PDB_2lox-17: 0.38, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.37, iPS_cells:PDB_1lox-21Puro-26: 0.37 |
| R63_w12GP_CTAAGTGGTGGATTTC-1 | Neurons:adrenal_medulla_cell_line | 0.12 | 557.87 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.35, Neuroepithelial_cell:ESC-derived: 0.31, Astrocyte:Embryonic_stem_cell-derived: 0.31, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.3, iPS_cells:iPS:minicircle-derived: 0.3, iPS_cells:PDB_2lox-5: 0.3, iPS_cells:PDB_1lox-17Puro-10: 0.3, Embryonic_stem_cells: 0.3, iPS_cells:PDB_1lox-17Puro-5: 0.3, iPS_cells:PDB_2lox-22: 0.3 |
| R30_w8.5_GAGTGAGAGCTAGAAT-1 | Neurons:adrenal_medulla_cell_line | 0.21 | 556.77 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.49, Neuroepithelial_cell:ESC-derived: 0.44, iPS_cells:PDB_1lox-17Puro-5: 0.42, iPS_cells:PDB_1lox-21Puro-20: 0.42, iPS_cells:PDB_1lox-17Puro-10: 0.42, iPS_cells:PDB_1lox-21Puro-26: 0.42, iPS_cells:PDB_2lox-22: 0.41, Astrocyte:Embryonic_stem_cell-derived: 0.41, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.41, iPS_cells:PDB_2lox-17: 0.41 |
| R53a_w9.5_GTGAGGAAGCACAAAT-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 556.26 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.39, Neuroepithelial_cell:ESC-derived: 0.34, Astrocyte:Embryonic_stem_cell-derived: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.32, iPS_cells:PDB_1lox-21Puro-20: 0.32, iPS_cells:PDB_1lox-21Puro-26: 0.32, Neurons:ES_cell-derived_neural_precursor: 0.32, iPS_cells:PDB_2lox-22: 0.32, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.32 |
| R53a_w9.5_GCACATATCGCACTCT-1 | Neurons:adrenal_medulla_cell_line | 0.18 | 548.18 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.46, Neuroepithelial_cell:ESC-derived: 0.42, Neurons:ES_cell-derived_neural_precursor: 0.4, iPS_cells:PDB_1lox-21Puro-20: 0.4, iPS_cells:PDB_1lox-17Puro-10: 0.4, iPS_cells:PDB_1lox-17Puro-5: 0.4, Astrocyte:Embryonic_stem_cell-derived: 0.4, iPS_cells:PDB_2lox-22: 0.4, iPS_cells:PDB_1lox-21Puro-26: 0.4, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.39 |
| R48a_w14.5_TCAAGTGAGGGCTAAC-1 | Neurons:adrenal_medulla_cell_line | 0.13 | 543.11 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.37, Neuroepithelial_cell:ESC-derived: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.32, Astrocyte:Embryonic_stem_cell-derived: 0.32, iPS_cells:PDB_1lox-17Puro-5: 0.32, iPS_cells:PDB_1lox-21Puro-20: 0.32, iPS_cells:PDB_2lox-22: 0.32, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.32, iPS_cells:PDB_1lox-21Puro-26: 0.32, Embryonic_stem_cells: 0.32 |
| R30_w8.5_TCTACATTCGTTCTAT-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 540.79 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.39, Neuroepithelial_cell:ESC-derived: 0.34, Astrocyte:Embryonic_stem_cell-derived: 0.33, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.32, Neurons:Schwann_cell: 0.32, Embryonic_stem_cells: 0.32, iPS_cells:PDB_1lox-21Puro-20: 0.32, iPS_cells:PDB_2lox-5: 0.32, iPS_cells:iPS:minicircle-derived: 0.31, iPS_cells:PDB_2lox-22: 0.31 |
| R53c_w9.5_AATCGACGTAGGATAT-1 | Neurons:adrenal_medulla_cell_line | 0.19 | 539.60 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.43, Neuroepithelial_cell:ESC-derived: 0.37, Neurons:ES_cell-derived_neural_precursor: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_2lox-22: 0.34, iPS_cells:PDB_2lox-5: 0.34 |
| R63_w12GP_TTCCTCTAGTAACGTA-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 535.00 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.37, Neuroepithelial_cell:ESC-derived: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.32, Astrocyte:Embryonic_stem_cell-derived: 0.31, iPS_cells:PDB_1lox-21Puro-20: 0.31, iPS_cells:PDB_1lox-21Puro-26: 0.31, iPS_cells:PDB_1lox-17Puro-5: 0.31, iPS_cells:PDB_2lox-5: 0.31, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.31, iPS_cells:PDB_2lox-21: 0.31 |
| R48c_w8.5_CATAGACAGCATTGAA-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 530.32 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.34, Neuroepithelial_cell:ESC-derived: 0.31, Astrocyte:Embryonic_stem_cell-derived: 0.29, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.28, iPS_cells:PDB_1lox-17Puro-5: 0.28, iPS_cells:PDB_1lox-17Puro-10: 0.28, iPS_cells:PDB_1lox-21Puro-20: 0.28, Embryonic_stem_cells: 0.28, iPS_cells:PDB_1lox-21Puro-26: 0.28, iPS_cells:PDB_2lox-5: 0.28 |
| R63_w12GP_AGGTCTAGTCCAAGAG-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 530.03 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.38, Neuroepithelial_cell:ESC-derived: 0.34, Astrocyte:Embryonic_stem_cell-derived: 0.32, iPS_cells:iPS:minicircle-derived: 0.32, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.32, iPS_cells:PDB_2lox-21: 0.32, iPS_cells:PDB_2lox-5: 0.32, iPS_cells:PDB_2lox-22: 0.32, iPS_cells:PDB_2lox-17: 0.32, Embryonic_stem_cells: 0.32 |
| R48b_w12_ACACTGACAAGACCTT-1 | Neurons:adrenal_medulla_cell_line | 0.18 | 524.37 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.43, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.36, iPS_cells:PDB_1lox-21Puro-20: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.35, iPS_cells:PDB_1lox-21Puro-26: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.35, iPS_cells:PDB_2lox-21: 0.34, iPS_cells:PDB_2lox-22: 0.34, Neurons:ES_cell-derived_neural_precursor: 0.34 |
Below shows the significant enrichments of this GEP for literature curated gene lists
This data was procured from existing single cell RNA-seq maps of neuroblastoma or related relevant data.
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by the P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
N Chromafin (Hanemaaijer)
Marker genes obtained from Supplementary Table SD of Hanemaaijer et al (PMID 33500353). The authors generated single-cell RNA-seq data (sort-seq, 2,229 cells total) from mouse adrenal glads at E13.5, E14.5, E17.5, E18.5, P1 and P5. These were marker genes that matched with a similar dataset generated by Furlan et al (PMID 28684471). This particular set of markers are for the Norepinepherine Chromaffin subcluster, which is part of the Adrenal Medulla cluster.:
Wilcoxon ranksum test P-value for gene set overrepresentation: 3.11e-09
Mean rank of genes in gene set: 2408.35
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| C1QL1 | 0.0126086 | 7 | GTEx | DepMap | Descartes | 4.41 | 675.65 |
| SCG5 | 0.0073173 | 26 | GTEx | DepMap | Descartes | 3.46 | 602.98 |
| SLC35D3 | 0.0071926 | 30 | GTEx | DepMap | Descartes | 0.38 | 33.13 |
| PTPRN | 0.0057951 | 51 | GTEx | DepMap | Descartes | 0.94 | 51.42 |
| GNAS | 0.0049285 | 78 | GTEx | DepMap | Descartes | 29.50 | 1678.68 |
| PCLO | 0.0036895 | 123 | GTEx | DepMap | Descartes | 1.53 | 22.77 |
| ADCYAP1R1 | 0.0034342 | 141 | GTEx | DepMap | Descartes | 0.32 | 14.27 |
| SCG3 | 0.0026224 | 215 | GTEx | DepMap | Descartes | 1.27 | 97.12 |
| CELF4 | 0.0025817 | 224 | GTEx | DepMap | Descartes | 1.09 | 80.93 |
| NAP1L5 | 0.0023978 | 264 | GTEx | DepMap | Descartes | 0.73 | 87.14 |
| CACNA2D1 | 0.0023471 | 273 | GTEx | DepMap | Descartes | 0.92 | 35.23 |
| SYN2 | 0.0009546 | 1018 | GTEx | DepMap | Descartes | 0.11 | 7.66 |
| PPFIA2 | 0.0008845 | 1135 | GTEx | DepMap | Descartes | 0.25 | 13.10 |
| SNAP25 | 0.0006311 | 1644 | GTEx | DepMap | Descartes | 1.15 | 127.18 |
| SLCO3A1 | 0.0004082 | 2531 | GTEx | DepMap | Descartes | 0.55 | 30.18 |
| LGR5 | 0.0000164 | 9198 | GTEx | DepMap | Descartes | 0.02 | 1.70 |
| CXCL14 | -0.0003485 | 23984 | GTEx | DepMap | Descartes | 0.01 | 1.30 |
Medulla (Hanemaaijer)
Marker genes obtained from Supplementary Table SD of Hanemaaijer et al (PMID 33500353). The authors generated single-cell RNA-seq data (sort-seq, 2,229 cells total) from mouse adrenal glads at E13.5, E14.5, E17.5, E18.5, P1 and P5. These were marker genes that matched with a similar dataset generated by Furlan et al (PMID 28684471). This particular set of markers are for genes defining the broad group labelled adrenal medulla, which included 7 subclusters (SCP, Bridge, Committed Progenitor, N Chromaffin, E Chromaffin, Neuroblast) - see UMAP on their Fig1B for cluster assignments.:
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.53e-08
Mean rank of genes in gene set: 2901.12
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CHGA | 0.0176475 | 1 | GTEx | DepMap | Descartes | 94.32 | 11651.42 |
| CHGB | 0.0163608 | 2 | GTEx | DepMap | Descartes | 49.98 | 4972.72 |
| TH | 0.0120792 | 8 | GTEx | DepMap | Descartes | 10.05 | 1450.43 |
| DBH | 0.0113835 | 10 | GTEx | DepMap | Descartes | 11.94 | 1169.41 |
| SLC18A1 | 0.0107091 | 12 | GTEx | DepMap | Descartes | 3.69 | 355.15 |
| PCSK1N | 0.0102016 | 13 | GTEx | DepMap | Descartes | 14.78 | 3168.28 |
| CYB561 | 0.0092274 | 16 | GTEx | DepMap | Descartes | 3.01 | 267.13 |
| DDC | 0.0055085 | 61 | GTEx | DepMap | Descartes | 2.09 | 261.04 |
| HAND2 | 0.0039656 | 113 | GTEx | DepMap | Descartes | 4.48 | 425.31 |
| PHOX2A | 0.0030858 | 166 | GTEx | DepMap | Descartes | 3.72 | 514.74 |
| EML5 | 0.0022589 | 288 | GTEx | DepMap | Descartes | 2.37 | 77.91 |
| GATA3 | 0.0021034 | 324 | GTEx | DepMap | Descartes | 2.27 | 214.28 |
| NNAT | 0.0008020 | 1265 | GTEx | DepMap | Descartes | 2.36 | 416.79 |
| UCHL1 | 0.0004498 | 2325 | GTEx | DepMap | Descartes | 2.94 | 397.03 |
| PHOX2B | 0.0003534 | 2879 | GTEx | DepMap | Descartes | 1.98 | 174.02 |
| DISP2 | -0.0000619 | 16383 | GTEx | DepMap | Descartes | 0.10 | 1.79 |
| MAP1B | -0.0006552 | 25453 | GTEx | DepMap | Descartes | 4.23 | 91.76 |
Committed Progenitor (Hanemaaijer)
Marker genes obtained from Supplementary Table SD of Hanemaaijer et al (PMID 33500353). The authors generated single-cell RNA-seq data (sort-seq, 2,229 cells total) from mouse adrenal glads at E13.5, E14.5, E17.5, E18.5, P1 and P5. These were marker genes that matched with a similar dataset generated by Furlan et al (PMID 28684471). This particular set of markers are for the Committed Progenitor subcluster (seems to mean progenitor of chromaffin cells), which is part of the Adrenal Medulla cluster.:
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.53e-06
Mean rank of genes in gene set: 4475.29
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| RGS4 | 0.0074418 | 25 | GTEx | DepMap | Descartes | 7.25 | 516.11 |
| MEG3 | 0.0070788 | 32 | GTEx | DepMap | Descartes | 69.04 | 1877.73 |
| DGKK | 0.0069569 | 33 | GTEx | DepMap | Descartes | 1.07 | 37.75 |
| RGS5 | 0.0068912 | 35 | GTEx | DepMap | Descartes | 15.06 | 667.31 |
| ZDBF2 | 0.0060973 | 47 | GTEx | DepMap | Descartes | 2.43 | 71.71 |
| SLC18A2 | 0.0057327 | 53 | GTEx | DepMap | Descartes | 1.48 | 96.41 |
| SYT1 | 0.0053281 | 65 | GTEx | DepMap | Descartes | 4.79 | 274.74 |
| FAM155A | 0.0050121 | 77 | GTEx | DepMap | Descartes | 0.71 | 21.69 |
| NDUFA4L2 | 0.0041498 | 104 | GTEx | DepMap | Descartes | 2.00 | 404.94 |
| UNC5C | 0.0034238 | 142 | GTEx | DepMap | Descartes | 1.20 | 37.32 |
| ATP6V1B2 | 0.0030603 | 168 | GTEx | DepMap | Descartes | 1.82 | 68.75 |
| C2CD4B | 0.0022325 | 292 | GTEx | DepMap | Descartes | 1.25 | 190.85 |
| PARM1 | 0.0009630 | 1006 | GTEx | DepMap | Descartes | 0.21 | 11.94 |
| SYTL4 | 0.0003292 | 3064 | GTEx | DepMap | Descartes | 0.70 | 38.71 |
| NTRK3 | -0.0001927 | 21683 | GTEx | DepMap | Descartes | 0.08 | 0.85 |
| AGTR2 | -0.0003134 | 23617 | GTEx | DepMap | Descartes | 0.04 | 2.17 |
| NRK | -0.0007925 | 25637 | GTEx | DepMap | Descartes | 0.24 | 4.31 |
Below shows ranks on this GEP for literature curated gene lists for large gene sets
These include those reported as mesenchymal/adrenergic by Van Groningen et al.
High ranks indicate this gene is a driver of this GEP (note these results are not ordered).
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
VanGroningen Adrenergic Genes
Adrenergic marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 3.98e-06
Mean rank of genes in gene set: 11166.78
Median rank of genes in gene set: 6002
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CHGA | 0.0176475 | 1 | GTEx | DepMap | Descartes | 94.32 | 11651.42 |
| CHGB | 0.0163608 | 2 | GTEx | DepMap | Descartes | 49.98 | 4972.72 |
| SCG2 | 0.0147153 | 3 | GTEx | DepMap | Descartes | 12.62 | 1285.25 |
| INSM1 | 0.0131589 | 6 | GTEx | DepMap | Descartes | 3.54 | 307.92 |
| TH | 0.0120792 | 8 | GTEx | DepMap | Descartes | 10.05 | 1450.43 |
| DBH | 0.0113835 | 10 | GTEx | DepMap | Descartes | 11.94 | 1169.41 |
| CHRNA3 | 0.0091114 | 17 | GTEx | DepMap | Descartes | 3.53 | 326.62 |
| QDPR | 0.0075939 | 24 | GTEx | DepMap | Descartes | 4.46 | 649.01 |
| RGS5 | 0.0068912 | 35 | GTEx | DepMap | Descartes | 15.06 | 667.31 |
| DLK1 | 0.0065554 | 40 | GTEx | DepMap | Descartes | 46.63 | 2296.78 |
| SYT4 | 0.0061707 | 45 | GTEx | DepMap | Descartes | 1.21 | 80.02 |
| GCH1 | 0.0055731 | 58 | GTEx | DepMap | Descartes | 1.41 | 112.35 |
| DDC | 0.0055085 | 61 | GTEx | DepMap | Descartes | 2.09 | 261.04 |
| SYT1 | 0.0053281 | 65 | GTEx | DepMap | Descartes | 4.79 | 274.74 |
| EEF1A2 | 0.0052812 | 70 | GTEx | DepMap | Descartes | 7.07 | 409.34 |
| PLPPR5 | 0.0051461 | 73 | GTEx | DepMap | Descartes | 0.65 | NA |
| FAM155A | 0.0050121 | 77 | GTEx | DepMap | Descartes | 0.71 | 21.69 |
| BEX2 | 0.0042412 | 97 | GTEx | DepMap | Descartes | 4.12 | 816.42 |
| BEX1 | 0.0042328 | 100 | GTEx | DepMap | Descartes | 8.60 | 2323.67 |
| GPR22 | 0.0040260 | 108 | GTEx | DepMap | Descartes | 1.20 | 96.79 |
| GRIA2 | 0.0040200 | 109 | GTEx | DepMap | Descartes | 2.35 | 127.40 |
| HAND2-AS1 | 0.0036133 | 129 | GTEx | DepMap | Descartes | 4.93 | NA |
| NPTX2 | 0.0035632 | 133 | GTEx | DepMap | Descartes | 0.48 | 43.53 |
| MIAT | 0.0035021 | 138 | GTEx | DepMap | Descartes | 2.29 | 82.98 |
| ADCYAP1R1 | 0.0034342 | 141 | GTEx | DepMap | Descartes | 0.32 | 14.27 |
| PHOX2A | 0.0030858 | 166 | GTEx | DepMap | Descartes | 3.72 | 514.74 |
| SHD | 0.0030673 | 167 | GTEx | DepMap | Descartes | 0.99 | 135.22 |
| ATP6V1B2 | 0.0030603 | 168 | GTEx | DepMap | Descartes | 1.82 | 68.75 |
| STXBP1 | 0.0030361 | 170 | GTEx | DepMap | Descartes | 1.55 | 86.24 |
| KLF13 | 0.0028691 | 185 | GTEx | DepMap | Descartes | 1.46 | 57.08 |
VanGroningen Mesenchymal Genes
Mesenchymal marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 18621.14
Median rank of genes in gene set: 23214
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| SERPINE2 | 0.0028270 | 189 | GTEx | DepMap | Descartes | 1.83 | 82.14 |
| ITM2C | 0.0026895 | 206 | GTEx | DepMap | Descartes | 3.46 | 397.67 |
| CD44 | 0.0026138 | 217 | GTEx | DepMap | Descartes | 2.07 | 95.74 |
| ATP1B1 | 0.0025275 | 234 | GTEx | DepMap | Descartes | 2.39 | 219.97 |
| HIST1H2AC | 0.0021373 | 310 | GTEx | DepMap | Descartes | 1.45 | NA |
| HSPA5 | 0.0020046 | 351 | GTEx | DepMap | Descartes | 5.23 | 327.04 |
| ROBO1 | 0.0019325 | 379 | GTEx | DepMap | Descartes | 0.87 | 32.88 |
| SHROOM3 | 0.0018331 | 406 | GTEx | DepMap | Descartes | 0.31 | 7.49 |
| KLF10 | 0.0015646 | 516 | GTEx | DepMap | Descartes | 0.95 | 68.37 |
| NPTN | 0.0015249 | 536 | GTEx | DepMap | Descartes | 1.22 | 103.87 |
| SDC2 | 0.0014626 | 565 | GTEx | DepMap | Descartes | 0.72 | 54.04 |
| SURF4 | 0.0014318 | 582 | GTEx | DepMap | Descartes | 1.68 | 139.22 |
| PTN | 0.0014221 | 590 | GTEx | DepMap | Descartes | 2.87 | 387.86 |
| EGR1 | 0.0013516 | 633 | GTEx | DepMap | Descartes | 3.78 | 304.77 |
| SSBP4 | 0.0013203 | 652 | GTEx | DepMap | Descartes | 2.10 | 279.38 |
| LMNA | 0.0013001 | 673 | GTEx | DepMap | Descartes | 1.72 | 126.83 |
| ATP6V0E1 | 0.0012898 | 684 | GTEx | DepMap | Descartes | 2.40 | 368.39 |
| DKK3 | 0.0012589 | 708 | GTEx | DepMap | Descartes | 1.06 | 27.40 |
| CRELD2 | 0.0012553 | 712 | GTEx | DepMap | Descartes | 0.69 | 59.54 |
| MBTPS1 | 0.0012103 | 748 | GTEx | DepMap | Descartes | 1.13 | 67.19 |
| MANF | 0.0011880 | 767 | GTEx | DepMap | Descartes | 1.57 | 218.13 |
| DMD | 0.0011702 | 791 | GTEx | DepMap | Descartes | 0.96 | 20.22 |
| SPRY4 | 0.0011116 | 836 | GTEx | DepMap | Descartes | 0.51 | 28.29 |
| FAM3C | 0.0011059 | 842 | GTEx | DepMap | Descartes | 0.97 | 91.88 |
| POLR2L | 0.0010700 | 876 | GTEx | DepMap | Descartes | 4.66 | 1078.84 |
| FAM43A | 0.0010520 | 900 | GTEx | DepMap | Descartes | 0.56 | 43.58 |
| NR3C1 | 0.0009938 | 960 | GTEx | DepMap | Descartes | 0.93 | 34.53 |
| ALDH1A3 | 0.0009350 | 1049 | GTEx | DepMap | Descartes | 0.10 | 9.19 |
| PTPRK | 0.0009063 | 1094 | GTEx | DepMap | Descartes | 0.44 | NA |
| STAT3 | 0.0008969 | 1111 | GTEx | DepMap | Descartes | 1.26 | 64.17 |
Descartes adrenocortical markers
Top 50 marker genes of adrenocortical cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/adrenocortical/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 20920.76
Median rank of genes in gene set: 24765.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| LINC00473 | 0.0024218 | 254 | GTEx | DepMap | Descartes | 0.67 | NA |
| JAKMIP2 | 0.0004762 | 2196 | GTEx | DepMap | Descartes | 0.55 | 14.67 |
| FRMD5 | 0.0003543 | 2874 | GTEx | DepMap | Descartes | 0.10 | 6.07 |
| NPC1 | 0.0003532 | 2883 | GTEx | DepMap | Descartes | 0.21 | 13.05 |
| TM7SF2 | 0.0003506 | 2906 | GTEx | DepMap | Descartes | 1.06 | 114.62 |
| FREM2 | 0.0001135 | 6034 | GTEx | DepMap | Descartes | 0.07 | 1.39 |
| SCARB1 | 0.0000845 | 6775 | GTEx | DepMap | Descartes | 1.19 | 56.12 |
| IGF1R | -0.0000099 | 10650 | GTEx | DepMap | Descartes | 0.72 | 16.17 |
| SGCZ | -0.0000878 | 18091 | GTEx | DepMap | Descartes | 0.00 | 0.07 |
| CLU | -0.0000949 | 18427 | GTEx | DepMap | Descartes | 0.92 | 79.51 |
| SLC2A14 | -0.0002058 | 21954 | GTEx | DepMap | Descartes | 0.02 | 0.68 |
| SCAP | -0.0002605 | 22934 | GTEx | DepMap | Descartes | 0.44 | 23.41 |
| ERN1 | -0.0002663 | 23026 | GTEx | DepMap | Descartes | 0.17 | 4.03 |
| SH3PXD2B | -0.0002777 | 23195 | GTEx | DepMap | Descartes | 0.11 | 3.40 |
| PDE10A | -0.0002880 | 23338 | GTEx | DepMap | Descartes | 0.09 | 2.18 |
| DNER | -0.0003330 | 23831 | GTEx | DepMap | Descartes | 0.08 | 5.83 |
| HMGCR | -0.0003368 | 23878 | GTEx | DepMap | Descartes | 0.70 | 34.25 |
| SLC16A9 | -0.0003409 | 23923 | GTEx | DepMap | Descartes | 0.22 | 13.56 |
| CYP17A1 | -0.0003754 | 24188 | GTEx | DepMap | Descartes | 1.26 | 55.39 |
| DHCR7 | -0.0004363 | 24638 | GTEx | DepMap | Descartes | 0.22 | 15.36 |
| PEG3 | -0.0004429 | 24676 | GTEx | DepMap | Descartes | 1.30 | NA |
| BAIAP2L1 | -0.0004468 | 24693 | GTEx | DepMap | Descartes | 0.04 | 2.04 |
| CYP11B1 | -0.0004550 | 24743 | GTEx | DepMap | Descartes | 0.34 | 16.53 |
| HMGCS1 | -0.0004631 | 24788 | GTEx | DepMap | Descartes | 0.87 | 34.79 |
| SLC1A2 | -0.0004780 | 24869 | GTEx | DepMap | Descartes | 0.04 | 0.75 |
| CYP21A2 | -0.0004799 | 24877 | GTEx | DepMap | Descartes | 0.38 | 22.78 |
| GSTA4 | -0.0004917 | 24928 | GTEx | DepMap | Descartes | 1.16 | 131.90 |
| SULT2A1 | -0.0005120 | 25027 | GTEx | DepMap | Descartes | 0.31 | 20.83 |
| MSMO1 | -0.0005242 | 25078 | GTEx | DepMap | Descartes | 0.72 | 60.54 |
| PAPSS2 | -0.0005473 | 25160 | GTEx | DepMap | Descartes | 0.09 | 4.00 |
Descartes chromaffin markers
Top 50 marker genes of chromaffin cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/chromaffin/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 22421.93
Median rank of genes in gene set: 25390
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| GAL | 0.0022638 | 287 | GTEx | DepMap | Descartes | 4.75 | 1408.79 |
| STMN2 | 0.0002681 | 3643 | GTEx | DepMap | Descartes | 14.80 | 1688.74 |
| EPHA6 | 0.0000586 | 7602 | GTEx | DepMap | Descartes | 0.04 | 2.98 |
| MAB21L1 | 0.0000313 | 8586 | GTEx | DepMap | Descartes | 0.76 | 68.01 |
| MAB21L2 | 0.0000213 | 9001 | GTEx | DepMap | Descartes | 0.36 | 35.03 |
| KCNB2 | -0.0000400 | 14442 | GTEx | DepMap | Descartes | 0.03 | 1.71 |
| EYA4 | -0.0000414 | 14612 | GTEx | DepMap | Descartes | 0.18 | 9.72 |
| NPY | -0.0001252 | 19691 | GTEx | DepMap | Descartes | 3.78 | 965.51 |
| TMEFF2 | -0.0002591 | 22915 | GTEx | DepMap | Descartes | 0.13 | 9.50 |
| ANKFN1 | -0.0002848 | 23296 | GTEx | DepMap | Descartes | 0.02 | 1.22 |
| SLC44A5 | -0.0003190 | 23671 | GTEx | DepMap | Descartes | 0.03 | 1.57 |
| EYA1 | -0.0003501 | 23998 | GTEx | DepMap | Descartes | 0.18 | 12.58 |
| RPH3A | -0.0003671 | 24109 | GTEx | DepMap | Descartes | 0.13 | 7.34 |
| CNTFR | -0.0003741 | 24176 | GTEx | DepMap | Descartes | 0.44 | 58.52 |
| RGMB | -0.0003989 | 24356 | GTEx | DepMap | Descartes | 0.32 | 17.79 |
| RYR2 | -0.0004990 | 24963 | GTEx | DepMap | Descartes | 0.04 | 0.51 |
| TUBB2A | -0.0005154 | 25038 | GTEx | DepMap | Descartes | 2.75 | 316.96 |
| FAT3 | -0.0005206 | 25056 | GTEx | DepMap | Descartes | 0.04 | 0.59 |
| TMEM132C | -0.0005560 | 25201 | GTEx | DepMap | Descartes | 0.09 | 5.31 |
| MLLT11 | -0.0005734 | 25259 | GTEx | DepMap | Descartes | 1.84 | 154.55 |
| REEP1 | -0.0005971 | 25332 | GTEx | DepMap | Descartes | 0.24 | 12.68 |
| HS3ST5 | -0.0006191 | 25376 | GTEx | DepMap | Descartes | 0.01 | 0.78 |
| BASP1 | -0.0006305 | 25404 | GTEx | DepMap | Descartes | 2.49 | 299.79 |
| MAP1B | -0.0006552 | 25453 | GTEx | DepMap | Descartes | 4.23 | 91.76 |
| ALK | -0.0006976 | 25524 | GTEx | DepMap | Descartes | 0.04 | 1.86 |
| CNKSR2 | -0.0007394 | 25579 | GTEx | DepMap | Descartes | 0.28 | 9.81 |
| NTRK1 | -0.0008044 | 25652 | GTEx | DepMap | Descartes | 0.06 | 3.12 |
| ISL1 | -0.0009108 | 25713 | GTEx | DepMap | Descartes | 0.98 | 94.61 |
| GREM1 | -0.0010743 | 25763 | GTEx | DepMap | Descartes | 0.06 | 1.02 |
| TUBB2B | -0.0010815 | 25766 | GTEx | DepMap | Descartes | 4.44 | 441.77 |
Descartes Vascular_endothelial markers
Top 50 marker genes of Vascular_endothelial cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/vascular_endothelial/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 24428.64
Median rank of genes in gene set: 25021.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CDH13 | 0.0000239 | 8890 | GTEx | DepMap | Descartes | 0.10 | 4.31 |
| NR5A2 | -0.0002445 | 22687 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| GALNT15 | -0.0002831 | 23266 | GTEx | DepMap | Descartes | 0.01 | NA |
| MYRIP | -0.0002844 | 23291 | GTEx | DepMap | Descartes | 0.01 | 0.32 |
| ESM1 | -0.0002994 | 23467 | GTEx | DepMap | Descartes | 0.02 | 1.63 |
| SHANK3 | -0.0003149 | 23630 | GTEx | DepMap | Descartes | 0.14 | 5.27 |
| SLCO2A1 | -0.0003629 | 24085 | GTEx | DepMap | Descartes | 0.01 | 0.93 |
| ID1 | -0.0003941 | 24320 | GTEx | DepMap | Descartes | 1.37 | 229.82 |
| DNASE1L3 | -0.0003942 | 24321 | GTEx | DepMap | Descartes | 0.05 | 5.49 |
| CRHBP | -0.0003993 | 24358 | GTEx | DepMap | Descartes | 0.03 | 3.53 |
| CHRM3 | -0.0004016 | 24374 | GTEx | DepMap | Descartes | 0.01 | 0.34 |
| APLNR | -0.0004063 | 24413 | GTEx | DepMap | Descartes | 0.02 | 1.21 |
| PODXL | -0.0004132 | 24476 | GTEx | DepMap | Descartes | 0.08 | 3.00 |
| FCGR2B | -0.0004209 | 24545 | GTEx | DepMap | Descartes | 0.03 | 1.62 |
| BTNL9 | -0.0004233 | 24565 | GTEx | DepMap | Descartes | 0.06 | 4.54 |
| NOTCH4 | -0.0004334 | 24628 | GTEx | DepMap | Descartes | 0.09 | 3.09 |
| SHE | -0.0004486 | 24703 | GTEx | DepMap | Descartes | 0.01 | 0.31 |
| CEACAM1 | -0.0004511 | 24721 | GTEx | DepMap | Descartes | 0.02 | 1.78 |
| FLT4 | -0.0004544 | 24741 | GTEx | DepMap | Descartes | 0.02 | 1.20 |
| IRX3 | -0.0004855 | 24901 | GTEx | DepMap | Descartes | 0.01 | 1.09 |
| CYP26B1 | -0.0004886 | 24914 | GTEx | DepMap | Descartes | 0.05 | 2.62 |
| TEK | -0.0005085 | 25013 | GTEx | DepMap | Descartes | 0.03 | 1.75 |
| NPR1 | -0.0005134 | 25030 | GTEx | DepMap | Descartes | 0.03 | 1.87 |
| TIE1 | -0.0005203 | 25055 | GTEx | DepMap | Descartes | 0.03 | 1.15 |
| RASIP1 | -0.0005210 | 25058 | GTEx | DepMap | Descartes | 0.08 | 4.62 |
| F8 | -0.0005263 | 25088 | GTEx | DepMap | Descartes | 0.04 | 1.14 |
| PTPRB | -0.0005266 | 25089 | GTEx | DepMap | Descartes | 0.04 | 0.83 |
| ROBO4 | -0.0005346 | 25113 | GTEx | DepMap | Descartes | 0.02 | 1.12 |
| EHD3 | -0.0005503 | 25175 | GTEx | DepMap | Descartes | 0.11 | 6.39 |
| MMRN2 | -0.0005664 | 25234 | GTEx | DepMap | Descartes | 0.03 | 2.64 |
Descartes stromal markers
Top 50 marker genes of stromal cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/stromal/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 21238.61
Median rank of genes in gene set: 22710
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ZNF385D | 0.0002050 | 4379 | GTEx | DepMap | Descartes | 0.07 | 1.62 |
| GAS2 | 0.0001759 | 4849 | GTEx | DepMap | Descartes | 0.05 | 5.60 |
| ABCC9 | 0.0001469 | 5374 | GTEx | DepMap | Descartes | 0.11 | 3.48 |
| GLI2 | 0.0000766 | 6999 | GTEx | DepMap | Descartes | 0.05 | 1.87 |
| DKK2 | -0.0000853 | 17957 | GTEx | DepMap | Descartes | 0.02 | 0.99 |
| CLDN11 | -0.0001074 | 18981 | GTEx | DepMap | Descartes | 0.16 | 11.61 |
| PCOLCE | -0.0001131 | 19216 | GTEx | DepMap | Descartes | 0.34 | 50.63 |
| ABCA6 | -0.0001135 | 19236 | GTEx | DepMap | Descartes | 0.01 | 0.48 |
| SCARA5 | -0.0001197 | 19478 | GTEx | DepMap | Descartes | 0.01 | 0.52 |
| ELN | -0.0001270 | 19753 | GTEx | DepMap | Descartes | 0.14 | 10.08 |
| CCDC102B | -0.0001273 | 19766 | GTEx | DepMap | Descartes | 0.06 | 5.45 |
| ITGA11 | -0.0001403 | 20227 | GTEx | DepMap | Descartes | 0.01 | 0.22 |
| ADAMTSL3 | -0.0001510 | 20605 | GTEx | DepMap | Descartes | 0.01 | 0.20 |
| BICC1 | -0.0001751 | 21259 | GTEx | DepMap | Descartes | 0.11 | 4.49 |
| FNDC1 | -0.0001795 | 21381 | GTEx | DepMap | Descartes | 0.01 | 0.34 |
| PAMR1 | -0.0001913 | 21663 | GTEx | DepMap | Descartes | 0.02 | 0.90 |
| POSTN | -0.0001960 | 21750 | GTEx | DepMap | Descartes | 0.27 | 16.04 |
| MXRA5 | -0.0001980 | 21785 | GTEx | DepMap | Descartes | 0.02 | 0.27 |
| LAMC3 | -0.0002044 | 21920 | GTEx | DepMap | Descartes | 0.01 | 0.35 |
| LUM | -0.0002062 | 21963 | GTEx | DepMap | Descartes | 0.04 | 1.72 |
| MGP | -0.0002202 | 22265 | GTEx | DepMap | Descartes | 0.27 | 19.03 |
| SFRP2 | -0.0002379 | 22577 | GTEx | DepMap | Descartes | 0.08 | 3.28 |
| LOX | -0.0002398 | 22606 | GTEx | DepMap | Descartes | 0.03 | 1.09 |
| PRICKLE1 | -0.0002418 | 22644 | GTEx | DepMap | Descartes | 0.07 | 2.69 |
| COL12A1 | -0.0002464 | 22710 | GTEx | DepMap | Descartes | 0.21 | 3.31 |
| ISLR | -0.0002755 | 23159 | GTEx | DepMap | Descartes | 0.10 | 7.51 |
| COL27A1 | -0.0002869 | 23323 | GTEx | DepMap | Descartes | 0.03 | 0.70 |
| HHIP | -0.0002979 | 23449 | GTEx | DepMap | Descartes | 0.09 | 1.15 |
| OGN | -0.0003048 | 23533 | GTEx | DepMap | Descartes | 0.14 | 7.83 |
| ACTA2 | -0.0003078 | 23565 | GTEx | DepMap | Descartes | 0.05 | 7.55 |
Descartes sympathoblasts markers
Top 50 marker genes of sympathoblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/sympathoblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 3.26e-29
Mean rank of genes in gene set: 382.64
Median rank of genes in gene set: 89.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CHGA | 0.0176475 | 1 | GTEx | DepMap | Descartes | 94.32 | 11651.42 |
| CHGB | 0.0163608 | 2 | GTEx | DepMap | Descartes | 49.98 | 4972.72 |
| SCG2 | 0.0147153 | 3 | GTEx | DepMap | Descartes | 12.62 | 1285.25 |
| PENK | 0.0132775 | 5 | GTEx | DepMap | Descartes | 8.46 | 1571.03 |
| INSM1 | 0.0131589 | 6 | GTEx | DepMap | Descartes | 3.54 | 307.92 |
| C1QL1 | 0.0126086 | 7 | GTEx | DepMap | Descartes | 4.41 | 675.65 |
| HTATSF1 | 0.0108788 | 11 | GTEx | DepMap | Descartes | 8.81 | 746.65 |
| SLC18A1 | 0.0107091 | 12 | GTEx | DepMap | Descartes | 3.69 | 355.15 |
| PCSK1N | 0.0102016 | 13 | GTEx | DepMap | Descartes | 14.78 | 3168.28 |
| PNMT | 0.0100336 | 14 | GTEx | DepMap | Descartes | 6.62 | 1387.62 |
| ARC | 0.0096922 | 15 | GTEx | DepMap | Descartes | 2.95 | 224.41 |
| SLC24A2 | 0.0086225 | 19 | GTEx | DepMap | Descartes | 1.25 | 33.30 |
| PCSK2 | 0.0072290 | 29 | GTEx | DepMap | Descartes | 1.40 | 82.06 |
| DGKK | 0.0069569 | 33 | GTEx | DepMap | Descartes | 1.07 | 37.75 |
| CCSER1 | 0.0068894 | 37 | GTEx | DepMap | Descartes | 1.38 | NA |
| ST18 | 0.0068163 | 38 | GTEx | DepMap | Descartes | 0.94 | 46.16 |
| CNTN3 | 0.0056991 | 55 | GTEx | DepMap | Descartes | 0.60 | 36.24 |
| FGF14 | 0.0056530 | 56 | GTEx | DepMap | Descartes | 1.26 | 26.60 |
| GCH1 | 0.0055731 | 58 | GTEx | DepMap | Descartes | 1.41 | 112.35 |
| SPOCK3 | 0.0052816 | 68 | GTEx | DepMap | Descartes | 0.79 | 62.78 |
| KSR2 | 0.0051765 | 72 | GTEx | DepMap | Descartes | 0.48 | 7.83 |
| FAM155A | 0.0050121 | 77 | GTEx | DepMap | Descartes | 0.71 | 21.69 |
| UNC80 | 0.0041988 | 102 | GTEx | DepMap | Descartes | 1.68 | 36.79 |
| LINC00632 | 0.0040019 | 110 | GTEx | DepMap | Descartes | 1.05 | NA |
| KCTD16 | 0.0035201 | 137 | GTEx | DepMap | Descartes | 0.42 | 8.16 |
| AGBL4 | 0.0034482 | 140 | GTEx | DepMap | Descartes | 0.36 | 22.77 |
| GALNTL6 | 0.0033449 | 148 | GTEx | DepMap | Descartes | 0.19 | 12.79 |
| PACRG | 0.0033355 | 149 | GTEx | DepMap | Descartes | 0.26 | 31.77 |
| TMEM130 | 0.0032380 | 159 | GTEx | DepMap | Descartes | 0.38 | 26.03 |
| MGAT4C | 0.0031190 | 164 | GTEx | DepMap | Descartes | 0.71 | 6.80 |
Descartes erythroblasts markers
Top 50 marker genes of erythroblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/erythroblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 23123.08
Median rank of genes in gene set: 24318
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| HECTD4 | 0.0002127 | 4288 | GTEx | DepMap | Descartes | 0.53 | NA |
| DENND4A | 0.0000503 | 7891 | GTEx | DepMap | Descartes | 0.27 | 8.89 |
| SLC25A21 | -0.0000849 | 17924 | GTEx | DepMap | Descartes | 0.02 | 1.06 |
| GCLC | -0.0001025 | 18772 | GTEx | DepMap | Descartes | 0.20 | 13.48 |
| RAPGEF2 | -0.0001380 | 20152 | GTEx | DepMap | Descartes | 0.37 | 11.22 |
| RHD | -0.0001718 | 21168 | GTEx | DepMap | Descartes | 0.01 | 0.34 |
| HBZ | -0.0001945 | 21725 | GTEx | DepMap | Descartes | 0.78 | 201.68 |
| RGS6 | -0.0002061 | 21959 | GTEx | DepMap | Descartes | 0.00 | 0.04 |
| TMCC2 | -0.0002171 | 22198 | GTEx | DepMap | Descartes | 0.14 | 5.29 |
| SPTB | -0.0002198 | 22244 | GTEx | DepMap | Descartes | 0.09 | 2.97 |
| ABCB10 | -0.0002568 | 22873 | GTEx | DepMap | Descartes | 0.14 | 9.56 |
| XPO7 | -0.0002618 | 22955 | GTEx | DepMap | Descartes | 0.24 | 12.90 |
| TMEM56 | -0.0002657 | 23012 | GTEx | DepMap | Descartes | 0.14 | NA |
| MICAL2 | -0.0002698 | 23077 | GTEx | DepMap | Descartes | 0.03 | 0.57 |
| RHCE | -0.0002711 | 23097 | GTEx | DepMap | Descartes | 0.05 | 6.02 |
| SNCA | -0.0002759 | 23171 | GTEx | DepMap | Descartes | 0.94 | 66.16 |
| TRAK2 | -0.0002821 | 23248 | GTEx | DepMap | Descartes | 0.20 | 6.74 |
| GYPE | -0.0003039 | 23523 | GTEx | DepMap | Descartes | 0.01 | 0.22 |
| EPB41 | -0.0003059 | 23547 | GTEx | DepMap | Descartes | 0.55 | 23.32 |
| TFR2 | -0.0003139 | 23622 | GTEx | DepMap | Descartes | 0.03 | 1.32 |
| SPTA1 | -0.0003163 | 23646 | GTEx | DepMap | Descartes | 0.05 | 1.15 |
| CR1L | -0.0003360 | 23859 | GTEx | DepMap | Descartes | 0.01 | 0.99 |
| ANK1 | -0.0003739 | 24173 | GTEx | DepMap | Descartes | 0.04 | 1.32 |
| SLC4A1 | -0.0003761 | 24196 | GTEx | DepMap | Descartes | 0.11 | 3.15 |
| RHAG | -0.0004088 | 24440 | GTEx | DepMap | Descartes | 0.04 | 2.05 |
| CPOX | -0.0004129 | 24472 | GTEx | DepMap | Descartes | 0.16 | 14.68 |
| TSPAN5 | -0.0004505 | 24717 | GTEx | DepMap | Descartes | 0.18 | 10.05 |
| FECH | -0.0004515 | 24725 | GTEx | DepMap | Descartes | 0.20 | 5.67 |
| EPB42 | -0.0004720 | 24838 | GTEx | DepMap | Descartes | 0.04 | 1.77 |
| HBG1 | -0.0004738 | 24850 | GTEx | DepMap | Descartes | 3.39 | 367.19 |
Descartes myeloid markers
Top 50 marker genes of myeloid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/myeloid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 21077.28
Median rank of genes in gene set: 22653.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| SFMBT2 | 0.0003220 | 3122 | GTEx | DepMap | Descartes | 0.19 | 6.29 |
| ITPR2 | 0.0001778 | 4820 | GTEx | DepMap | Descartes | 0.43 | 9.76 |
| CST3 | 0.0001369 | 5563 | GTEx | DepMap | Descartes | 2.41 | 161.29 |
| IFNGR1 | 0.0000513 | 7847 | GTEx | DepMap | Descartes | 0.36 | 36.28 |
| HRH1 | -0.0000285 | 13093 | GTEx | DepMap | Descartes | 0.13 | 5.59 |
| MS4A4E | -0.0000597 | 16221 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| RBPJ | -0.0000852 | 17943 | GTEx | DepMap | Descartes | 1.34 | 54.60 |
| SPP1 | -0.0001288 | 19833 | GTEx | DepMap | Descartes | 0.03 | 3.32 |
| CD163L1 | -0.0001481 | 20515 | GTEx | DepMap | Descartes | 0.01 | 0.41 |
| HCK | -0.0001505 | 20588 | GTEx | DepMap | Descartes | 0.01 | 0.75 |
| LGMN | -0.0001693 | 21112 | GTEx | DepMap | Descartes | 0.37 | 43.28 |
| CD14 | -0.0001764 | 21305 | GTEx | DepMap | Descartes | 0.11 | 10.62 |
| FGD2 | -0.0001780 | 21341 | GTEx | DepMap | Descartes | 0.01 | 0.45 |
| ATP8B4 | -0.0001937 | 21700 | GTEx | DepMap | Descartes | 0.01 | 0.24 |
| MSR1 | -0.0001956 | 21743 | GTEx | DepMap | Descartes | 0.01 | 0.43 |
| MS4A4A | -0.0002050 | 21928 | GTEx | DepMap | Descartes | 0.02 | 1.53 |
| ADAP2 | -0.0002052 | 21933 | GTEx | DepMap | Descartes | 0.03 | 2.37 |
| VSIG4 | -0.0002090 | 22028 | GTEx | DepMap | Descartes | 0.01 | 1.21 |
| MPEG1 | -0.0002106 | 22064 | GTEx | DepMap | Descartes | 0.02 | 0.69 |
| SLCO2B1 | -0.0002121 | 22089 | GTEx | DepMap | Descartes | 0.01 | 0.26 |
| HLA-DPA1 | -0.0002197 | 22241 | GTEx | DepMap | Descartes | 0.06 | 2.39 |
| CTSD | -0.0002206 | 22276 | GTEx | DepMap | Descartes | 0.87 | 106.13 |
| MARCH1 | -0.0002218 | 22299 | GTEx | DepMap | Descartes | 0.02 | NA |
| CD163 | -0.0002296 | 22439 | GTEx | DepMap | Descartes | 0.04 | 1.81 |
| MS4A7 | -0.0002422 | 22650 | GTEx | DepMap | Descartes | 0.03 | 1.78 |
| C1QB | -0.0002427 | 22657 | GTEx | DepMap | Descartes | 0.06 | 13.55 |
| CYBB | -0.0002436 | 22671 | GTEx | DepMap | Descartes | 0.04 | 2.18 |
| C1QC | -0.0002464 | 22712 | GTEx | DepMap | Descartes | 0.04 | 4.85 |
| CTSS | -0.0002476 | 22731 | GTEx | DepMap | Descartes | 0.02 | 1.03 |
| C1QA | -0.0002568 | 22874 | GTEx | DepMap | Descartes | 0.09 | 17.70 |
Descartes Schwann markers
Top 50 marker genes of Schwann cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/schwann/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.98e-01
Mean rank of genes in gene set: 16043.84
Median rank of genes in gene set: 22504
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| NRXN3 | 0.0020236 | 341 | GTEx | DepMap | Descartes | 0.34 | 11.36 |
| SORCS1 | 0.0019436 | 373 | GTEx | DepMap | Descartes | 0.30 | 11.48 |
| EGFLAM | 0.0015728 | 511 | GTEx | DepMap | Descartes | 0.36 | 21.57 |
| NRXN1 | 0.0015625 | 517 | GTEx | DepMap | Descartes | 1.23 | 37.60 |
| PTN | 0.0014221 | 590 | GTEx | DepMap | Descartes | 2.87 | 387.86 |
| GRIK3 | 0.0011104 | 837 | GTEx | DepMap | Descartes | 0.13 | 3.79 |
| LRRTM4 | 0.0007224 | 1437 | GTEx | DepMap | Descartes | 0.07 | 5.13 |
| GAS7 | 0.0006573 | 1588 | GTEx | DepMap | Descartes | 0.28 | 9.30 |
| PAG1 | 0.0005474 | 1923 | GTEx | DepMap | Descartes | 0.90 | 23.77 |
| GFRA3 | 0.0002051 | 4376 | GTEx | DepMap | Descartes | 0.43 | 56.71 |
| COL18A1 | 0.0001772 | 4830 | GTEx | DepMap | Descartes | 0.63 | 29.16 |
| IL1RAPL2 | 0.0001461 | 5394 | GTEx | DepMap | Descartes | 0.01 | 1.52 |
| MDGA2 | 0.0000938 | 6522 | GTEx | DepMap | Descartes | 0.02 | 0.99 |
| SCN7A | 0.0000917 | 6578 | GTEx | DepMap | Descartes | 0.15 | 4.48 |
| SOX5 | 0.0000915 | 6588 | GTEx | DepMap | Descartes | 0.22 | 7.46 |
| FIGN | 0.0000496 | 7914 | GTEx | DepMap | Descartes | 0.17 | 4.94 |
| DST | 0.0000399 | 8261 | GTEx | DepMap | Descartes | 2.94 | 37.77 |
| NLGN4X | 0.0000214 | 8995 | GTEx | DepMap | Descartes | 0.24 | 12.43 |
| STARD13 | -0.0000191 | 11801 | GTEx | DepMap | Descartes | 0.14 | 5.79 |
| TRPM3 | -0.0000758 | 17356 | GTEx | DepMap | Descartes | 0.09 | 2.00 |
| ERBB4 | -0.0000946 | 18415 | GTEx | DepMap | Descartes | 0.03 | 1.26 |
| COL25A1 | -0.0001312 | 19925 | GTEx | DepMap | Descartes | 0.08 | 2.49 |
| ZNF536 | -0.0002251 | 22364 | GTEx | DepMap | Descartes | 0.11 | 5.75 |
| SOX10 | -0.0002315 | 22471 | GTEx | DepMap | Descartes | 0.02 | 1.56 |
| CDH19 | -0.0002336 | 22504 | GTEx | DepMap | Descartes | 0.02 | 0.90 |
| IL1RAPL1 | -0.0002346 | 22519 | GTEx | DepMap | Descartes | 0.00 | 0.37 |
| LAMC1 | -0.0002504 | 22777 | GTEx | DepMap | Descartes | 0.32 | 10.97 |
| MPZ | -0.0002507 | 22785 | GTEx | DepMap | Descartes | 0.05 | 4.97 |
| ADAMTS5 | -0.0002637 | 22984 | GTEx | DepMap | Descartes | 0.02 | 0.35 |
| PLCE1 | -0.0002664 | 23027 | GTEx | DepMap | Descartes | 0.06 | 0.92 |
Descartes Megakaryocytes markers
Top 50 marker genes of Megakaryocytes cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/megakaryocytes/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 20545.02
Median rank of genes in gene set: 22740
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CD9 | 0.0011898 | 764 | GTEx | DepMap | Descartes | 1.74 | 229.20 |
| RAB27B | 0.0009530 | 1021 | GTEx | DepMap | Descartes | 0.41 | 15.12 |
| ACTN1 | 0.0007029 | 1474 | GTEx | DepMap | Descartes | 0.61 | 33.02 |
| ANGPT1 | 0.0001513 | 5285 | GTEx | DepMap | Descartes | 0.22 | 13.73 |
| SLC2A3 | 0.0000498 | 7906 | GTEx | DepMap | Descartes | 1.11 | 64.28 |
| PDE3A | -0.0000491 | 15343 | GTEx | DepMap | Descartes | 0.17 | 6.06 |
| PPBP | -0.0000710 | 17050 | GTEx | DepMap | Descartes | 0.01 | 1.33 |
| PF4 | -0.0000726 | 17142 | GTEx | DepMap | Descartes | 0.01 | 2.17 |
| ITGA2B | -0.0000726 | 17144 | GTEx | DepMap | Descartes | 0.01 | 1.16 |
| ITGB3 | -0.0000805 | 17634 | GTEx | DepMap | Descartes | 0.00 | 0.06 |
| SLC24A3 | -0.0001027 | 18790 | GTEx | DepMap | Descartes | 0.01 | 0.79 |
| GP1BA | -0.0001036 | 18827 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TUBB1 | -0.0001144 | 19271 | GTEx | DepMap | Descartes | 0.04 | 4.23 |
| STON2 | -0.0001173 | 19387 | GTEx | DepMap | Descartes | 0.11 | 6.49 |
| GP9 | -0.0001362 | 20096 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TRPC6 | -0.0001597 | 20864 | GTEx | DepMap | Descartes | 0.00 | 0.23 |
| P2RX1 | -0.0001710 | 21150 | GTEx | DepMap | Descartes | 0.00 | 0.07 |
| CD84 | -0.0001800 | 21399 | GTEx | DepMap | Descartes | 0.00 | 0.04 |
| MCTP1 | -0.0001933 | 21694 | GTEx | DepMap | Descartes | 0.00 | 0.08 |
| PLEK | -0.0001979 | 21783 | GTEx | DepMap | Descartes | 0.01 | 1.18 |
| BIN2 | -0.0002097 | 22039 | GTEx | DepMap | Descartes | 0.01 | 1.28 |
| FERMT3 | -0.0002346 | 22518 | GTEx | DepMap | Descartes | 0.01 | 1.47 |
| SPN | -0.0002407 | 22625 | GTEx | DepMap | Descartes | 0.01 | 0.29 |
| MED12L | -0.0002472 | 22725 | GTEx | DepMap | Descartes | 0.11 | 3.20 |
| MYLK | -0.0002482 | 22740 | GTEx | DepMap | Descartes | 0.13 | 3.66 |
| THBS1 | -0.0002795 | 23221 | GTEx | DepMap | Descartes | 0.04 | 1.07 |
| HIPK2 | -0.0002801 | 23226 | GTEx | DepMap | Descartes | 0.43 | 7.25 |
| UBASH3B | -0.0002837 | 23277 | GTEx | DepMap | Descartes | 0.01 | 0.50 |
| ARHGAP6 | -0.0002983 | 23457 | GTEx | DepMap | Descartes | 0.01 | 0.31 |
| PSTPIP2 | -0.0003017 | 23500 | GTEx | DepMap | Descartes | 0.02 | 1.30 |
Descartes Lyphoid markers
Top 50 marker genes of Lyphoid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/lymphoid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 19213.94
Median rank of genes in gene set: 23690
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CD44 | 0.0026138 | 217 | GTEx | DepMap | Descartes | 2.07 | 95.74 |
| SCML4 | 0.0018243 | 411 | GTEx | DepMap | Descartes | 0.20 | 12.91 |
| RAP1GAP2 | 0.0012532 | 715 | GTEx | DepMap | Descartes | 0.23 | 9.41 |
| DOCK10 | 0.0007133 | 1455 | GTEx | DepMap | Descartes | 0.27 | 11.95 |
| SORL1 | 0.0006295 | 1647 | GTEx | DepMap | Descartes | 0.42 | 12.23 |
| ABLIM1 | 0.0001279 | 5725 | GTEx | DepMap | Descartes | 0.35 | 12.80 |
| CELF2 | 0.0001104 | 6123 | GTEx | DepMap | Descartes | 0.26 | 8.19 |
| EVL | 0.0000952 | 6489 | GTEx | DepMap | Descartes | 1.20 | 85.44 |
| ANKRD44 | 0.0000487 | 7949 | GTEx | DepMap | Descartes | 0.29 | 10.55 |
| WIPF1 | 0.0000020 | 9823 | GTEx | DepMap | Descartes | 0.17 | 9.83 |
| PITPNC1 | -0.0000155 | 11288 | GTEx | DepMap | Descartes | 0.23 | 8.42 |
| LCP1 | -0.0000304 | 13349 | GTEx | DepMap | Descartes | 0.12 | 7.81 |
| STK39 | -0.0000410 | 14572 | GTEx | DepMap | Descartes | 0.27 | 19.40 |
| LINC00299 | -0.0001225 | 19589 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CCL5 | -0.0001414 | 20262 | GTEx | DepMap | Descartes | 0.02 | 3.43 |
| SKAP1 | -0.0001493 | 20553 | GTEx | DepMap | Descartes | 0.02 | 1.67 |
| FYN | -0.0001630 | 20956 | GTEx | DepMap | Descartes | 0.71 | 49.11 |
| NKG7 | -0.0001790 | 21363 | GTEx | DepMap | Descartes | 0.02 | 8.88 |
| ARHGAP15 | -0.0002442 | 22681 | GTEx | DepMap | Descartes | 0.01 | 1.04 |
| NCALD | -0.0002605 | 22933 | GTEx | DepMap | Descartes | 0.18 | 9.63 |
| SAMD3 | -0.0002660 | 23013 | GTEx | DepMap | Descartes | 0.01 | 0.49 |
| PTPRC | -0.0003043 | 23526 | GTEx | DepMap | Descartes | 0.03 | 1.19 |
| PLEKHA2 | -0.0003154 | 23634 | GTEx | DepMap | Descartes | 0.04 | 1.99 |
| IKZF1 | -0.0003194 | 23679 | GTEx | DepMap | Descartes | 0.01 | 0.26 |
| MCTP2 | -0.0003210 | 23701 | GTEx | DepMap | Descartes | 0.02 | 0.33 |
| ITPKB | -0.0003273 | 23766 | GTEx | DepMap | Descartes | 0.02 | 0.62 |
| PDE3B | -0.0003478 | 23978 | GTEx | DepMap | Descartes | 0.07 | 2.89 |
| RCSD1 | -0.0003521 | 24015 | GTEx | DepMap | Descartes | 0.01 | 0.39 |
| HLA-C | -0.0003628 | 24084 | GTEx | DepMap | Descartes | 0.63 | 73.06 |
| PRKCH | -0.0004041 | 24391 | GTEx | DepMap | Descartes | 0.02 | 0.85 |
Below shows the ranks on this GEP for immune marker gene lists curated by CellTypist (https://www.celltypist.org/encyclopedia).
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Granulocytes: Neutrophils (model markers)
the most abundant type of granulocytes that contains distinctive cytoplasmic granules and forms an essential part of the innate immune system:
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.91e-02
Mean rank of genes in gene set: 4760.33
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| GATA3-AS1 | 0.0019434 | 374 | GTEx | DepMap | Descartes | 0.58 | NA |
| OR2A25 | 0.0000882 | 6657 | GTEx | DepMap | Descartes | 0.00 | 0.34 |
| LINC01709 | 0.0000694 | 7250 | GTEx | DepMap | Descartes | 0.00 | NA |
No detectable expression in this dataset: IGHV5-78
Macrophages: Hofbauer cells (model markers)
primitive placental resident macrophages with granules and vacuoles found in placenta particularly during early pregnancy:
Wilcoxon ranksum test P-value for gene set overrepresentation: 4.30e-02
Mean rank of genes in gene set: 8641.44
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| GATA3-AS1 | 0.0019434 | 374 | GTEx | DepMap | Descartes | 0.58 | NA |
| CGA | 0.0007623 | 1355 | GTEx | DepMap | Descartes | 0.06 | 18.21 |
| PANX2 | 0.0006256 | 1660 | GTEx | DepMap | Descartes | 0.09 | 8.48 |
| PAGE4 | 0.0001826 | 4742 | GTEx | DepMap | Descartes | 0.02 | 1.68 |
| ZFHX4-AS1 | 0.0000867 | 6709 | GTEx | DepMap | Descartes | 0.01 | NA |
| KIAA0087 | 0.0000133 | 9319 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| XAGE3 | -0.0000268 | 12872 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| GAPLINC | -0.0000484 | 15285 | GTEx | DepMap | Descartes | 0.00 | NA |
| LYVE1 | -0.0006577 | 25457 | GTEx | DepMap | Descartes | 0.17 | 10.44 |
Plasma cells: Plasma cells (model markers)
B-lymphocyte white blood cells capable of secreting large quantities of immunoglobulins or antibodies:
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.11e-02
Mean rank of genes in gene set: 9149.86
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ST18 | 0.0068163 | 38 | GTEx | DepMap | Descartes | 0.94 | 46.16 |
| TGFBR3L | 0.0019663 | 366 | GTEx | DepMap | Descartes | 0.14 | NA |
| XBP1 | 0.0005927 | 1766 | GTEx | DepMap | Descartes | 0.84 | 84.88 |
| IGKC | -0.0000355 | 13944 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| JCHAIN | -0.0000416 | 14631 | GTEx | DepMap | Descartes | 0.00 | NA |
| MZB1 | -0.0000620 | 16395 | GTEx | DepMap | Descartes | 0.00 | NA |
| C11orf72 | -0.0000690 | 16909 | GTEx | DepMap | Descartes | 0.00 | 0.00 |