Program: 4. Macrophage.
Program description and justification of annotation: 4.
Submit a comment on this gene expression program’s interpretation: CLICK
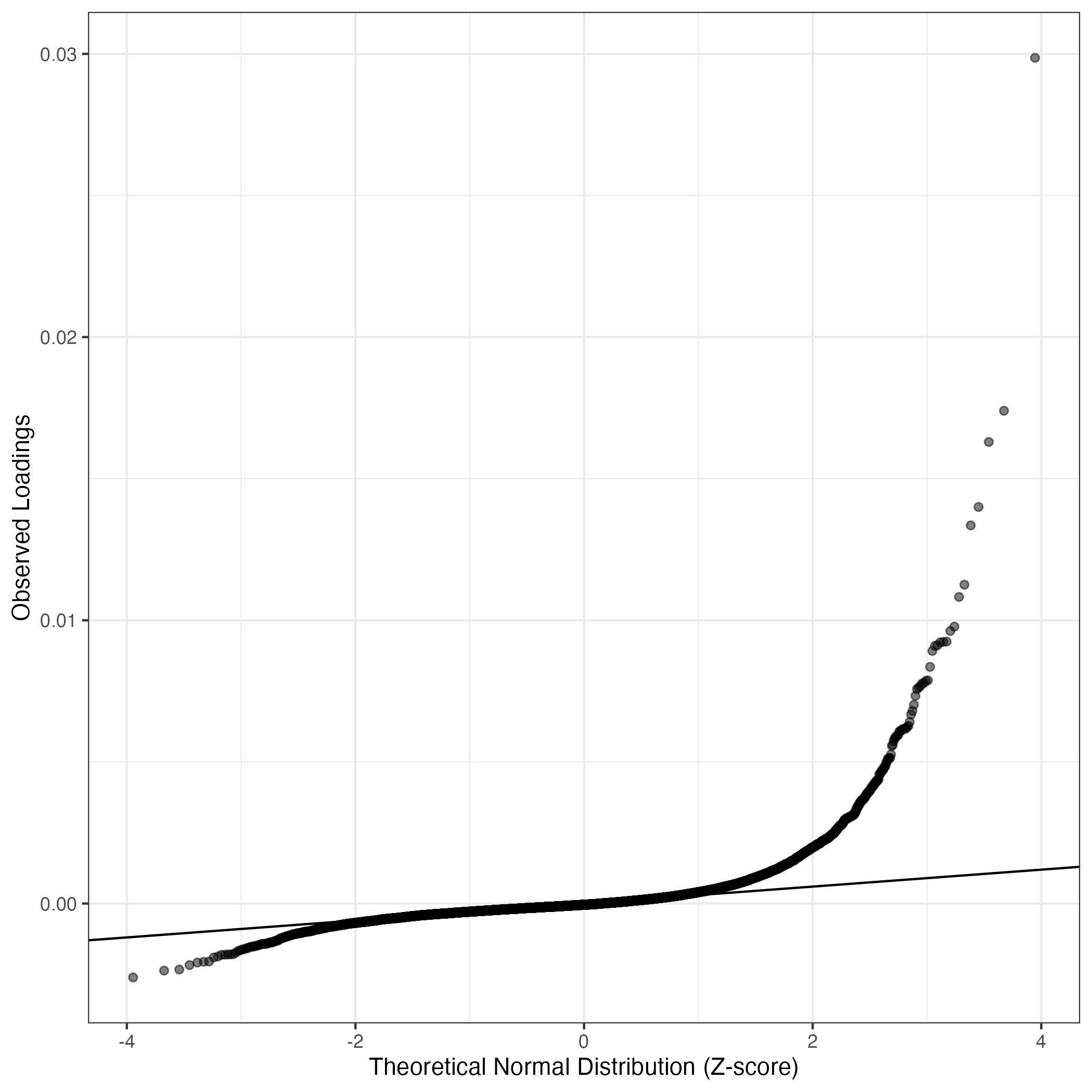
QQ-plot of gene loadings, averaged over both independent splits of the data
This plot highlights the relative contribution of each gene to the GEP
Top genes driving this program.
Note: Decartes website is buggy, try refreshing. Also, Decartes fetal adrenal data have been collected at specific time points (89-122 days), all possible cell types of interest may not be represented, do not overinterpret.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
| Gene | Loading | Gene.Name | GTEx | DepMap | Descartes | Mean.Counts | Mean.Tpm | |
|---|---|---|---|---|---|---|---|---|
| 1 | SPP1 | 0.0298595 | secreted phosphoprotein 1 | GTEx | DepMap | Descartes | 11.14 | 16904.96 |
| 2 | MMP12 | 0.0173984 | matrix metallopeptidase 12 | GTEx | DepMap | Descartes | 0.43 | 694.16 |
| 3 | APOE | 0.0162972 | apolipoprotein E | GTEx | DepMap | Descartes | 2.06 | 4173.35 |
| 4 | APOC1 | 0.0140010 | apolipoprotein C1 | GTEx | DepMap | Descartes | 1.35 | 4965.71 |
| 5 | CTSD | 0.0133510 | cathepsin D | GTEx | DepMap | Descartes | 1.66 | 2008.08 |
| 6 | ACP5 | 0.0112574 | acid phosphatase 5, tartrate resistant | GTEx | DepMap | Descartes | 0.34 | 531.48 |
| 7 | MITF | 0.0108242 | melanocyte inducing transcription factor | GTEx | DepMap | Descartes | 0.57 | 282.58 |
| 8 | CTSB | 0.0097785 | cathepsin B | GTEx | DepMap | Descartes | 1.21 | 829.79 |
| 9 | CTSZ | 0.0096236 | cathepsin Z | GTEx | DepMap | Descartes | 0.64 | 937.30 |
| 10 | NEAT1 | 0.0092469 | nuclear paraspeckle assembly transcript 1 | GTEx | DepMap | Descartes | 6.58 | 596.42 |
| 11 | SLC2A3 | 0.0092391 | solute carrier family 2 member 3 | GTEx | DepMap | Descartes | 0.64 | 323.63 |
| 12 | SLC11A1 | 0.0092208 | solute carrier family 11 member 1 | GTEx | DepMap | Descartes | 0.32 | 150.29 |
| 13 | HMOX1 | 0.0091138 | heme oxygenase 1 | GTEx | DepMap | Descartes | 0.30 | 466.62 |
| 14 | ITGAX | 0.0090952 | integrin subunit alpha X | GTEx | DepMap | Descartes | 0.37 | 192.72 |
| 15 | IL1RN | 0.0089180 | interleukin 1 receptor antagonist | GTEx | DepMap | Descartes | 0.18 | 226.90 |
| 16 | GPNMB | 0.0083548 | glycoprotein nmb | GTEx | DepMap | Descartes | 0.45 | 394.84 |
| 17 | CAPG | 0.0078786 | capping actin protein, gelsolin like | GTEx | DepMap | Descartes | 0.18 | 316.73 |
| 18 | SLC16A10 | 0.0078610 | solute carrier family 16 member 10 | GTEx | DepMap | Descartes | 0.70 | 198.47 |
| 19 | LAPTM5 | 0.0077848 | lysosomal protein transmembrane 5 | GTEx | DepMap | Descartes | 0.36 | 379.38 |
| 20 | PLAUR | 0.0077670 | plasminogen activator, urokinase receptor | GTEx | DepMap | Descartes | 0.39 | 658.70 |
| 21 | CD109 | 0.0076756 | CD109 molecule | GTEx | DepMap | Descartes | 0.18 | 43.88 |
| 22 | TYROBP | 0.0076193 | transmembrane immune signaling adaptor TYROBP | GTEx | DepMap | Descartes | 0.28 | 1005.51 |
| 23 | TM4SF19 | 0.0075697 | transmembrane 4 L six family member 19 | GTEx | DepMap | Descartes | 0.01 | 11.25 |
| 24 | MMP19 | 0.0073309 | matrix metallopeptidase 19 | GTEx | DepMap | Descartes | 0.24 | 173.22 |
| 25 | TRPM3 | 0.0070133 | transient receptor potential cation channel subfamily M member 3 | GTEx | DepMap | Descartes | 0.95 | 166.63 |
| 26 | CHST11 | 0.0068036 | carbohydrate sulfotransferase 11 | GTEx | DepMap | Descartes | 1.03 | 431.72 |
| 27 | PSAP | 0.0066719 | prosaposin | GTEx | DepMap | Descartes | 1.51 | 1173.42 |
| 28 | CD14 | 0.0064117 | CD14 molecule | GTEx | DepMap | Descartes | 0.23 | 346.68 |
| 29 | RGS1 | 0.0062721 | regulator of G protein signaling 1 | GTEx | DepMap | Descartes | 0.28 | 263.52 |
| 30 | TFRC | 0.0062348 | transferrin receptor | GTEx | DepMap | Descartes | 0.74 | 396.03 |
| 31 | ARHGAP26 | 0.0061790 | Rho GTPase activating protein 26 | GTEx | DepMap | Descartes | 2.36 | 512.95 |
| 32 | TGFBI | 0.0061742 | transforming growth factor beta induced | GTEx | DepMap | Descartes | 0.32 | 164.77 |
| 33 | ZEB2 | 0.0061579 | zinc finger E-box binding homeobox 2 | GTEx | DepMap | Descartes | 1.67 | 413.08 |
| 34 | CSTB | 0.0061527 | cystatin B | GTEx | DepMap | Descartes | 0.50 | NA |
| 35 | ITGB2 | 0.0061108 | integrin subunit beta 2 | GTEx | DepMap | Descartes | 0.23 | 102.67 |
| 36 | SLC16A3 | 0.0060900 | solute carrier family 16 member 3 | GTEx | DepMap | Descartes | 0.26 | 121.21 |
| 37 | RAB31 | 0.0060777 | RAB31, member RAS oncogene family | GTEx | DepMap | Descartes | 0.36 | 219.44 |
| 38 | CD68 | 0.0059503 | CD68 molecule | GTEx | DepMap | Descartes | 0.08 | 96.09 |
| 39 | S100A10 | 0.0059398 | S100 calcium binding protein A10 | GTEx | DepMap | Descartes | 0.18 | 593.98 |
| 40 | SRGN | 0.0059010 | serglycin | GTEx | DepMap | Descartes | 0.49 | 949.73 |
| 41 | PAM | 0.0058616 | peptidylglycine alpha-amidating monooxygenase | GTEx | DepMap | Descartes | 2.04 | 768.86 |
| 42 | S100A4 | 0.0058178 | S100 calcium binding protein A4 | GTEx | DepMap | Descartes | 0.22 | 700.92 |
| 43 | FCER1G | 0.0057312 | Fc epsilon receptor Ig | GTEx | DepMap | Descartes | 0.17 | 649.08 |
| 44 | RAB20 | 0.0055960 | RAB20, member RAS oncogene family | GTEx | DepMap | Descartes | 0.16 | 260.03 |
| 45 | GNAS | 0.0055629 | GNAS complex locus | GTEx | DepMap | Descartes | 4.19 | 2001.95 |
| 46 | HIF1A | 0.0052630 | hypoxia inducible factor 1 subunit alpha | GTEx | DepMap | Descartes | 1.00 | 580.88 |
| 47 | RREB1 | 0.0051386 | ras responsive element binding protein 1 | GTEx | DepMap | Descartes | 0.29 | 64.81 |
| 48 | ELL2 | 0.0051379 | elongation factor for RNA polymerase II 2 | GTEx | DepMap | Descartes | 0.65 | 228.79 |
| 49 | B2M | 0.0051099 | beta-2-microglobulin | GTEx | DepMap | Descartes | 2.15 | 1602.43 |
| 50 | HK2 | 0.0051072 | hexokinase 2 | GTEx | DepMap | Descartes | 0.16 | 70.90 |
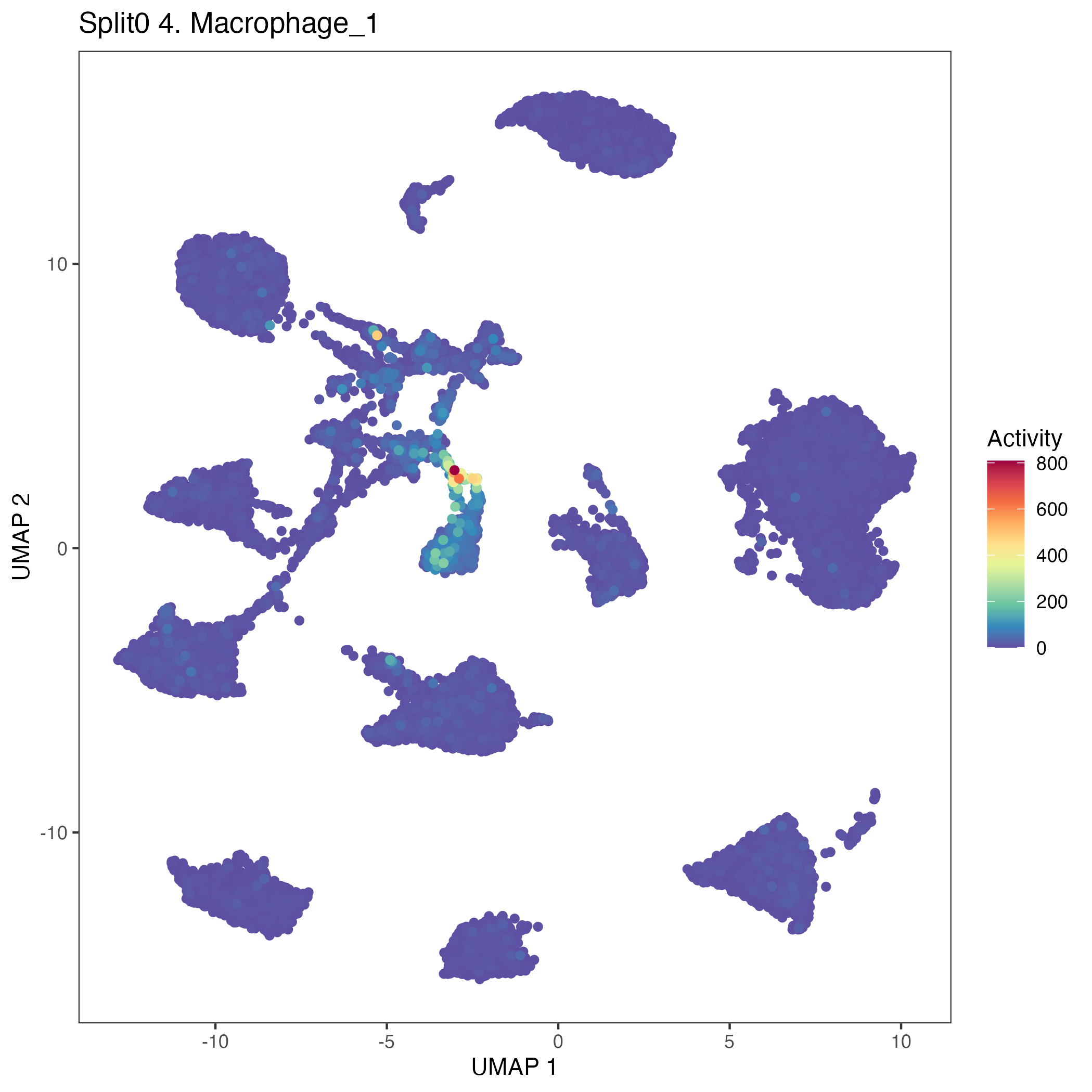
UMAP plots showing activity of gene expression program identified in community:4. Macrophage
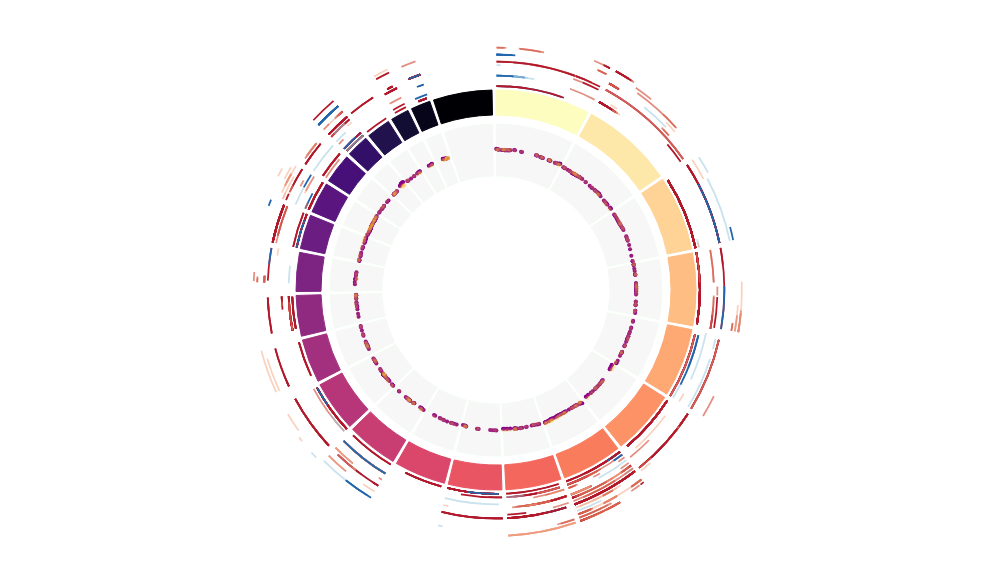
CNV Data procured from inferCNV.
Outer tracks are putative CNV regions (gains = red, losses = blue) for each patient
Inner track is expression data representing:
The top cells expressing this GEP (purple)
Random cells (n =50) from the reference set used in inferCNV (orange)
Gene set Enrichments for this program, caculated from top 50 genes
mSigDB Cell Types Gene Set:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| DURANTE_ADULT_OLFACTORY_NEUROEPITHELIUM_MACROPHAGES | 1.14e-14 | 50.30 | 22.29 | 4.78e-13 | 7.65e-12 | 11APOE, APOC1, LAPTM5, PLAUR, TYROBP, CD14, RGS1, ITGB2, CD68, SRGN, FCER1G |
81 |
| DURANTE_ADULT_OLFACTORY_NEUROEPITHELIUM_DENDRITIC_CELLS | 4.80e-16 | 42.17 | 19.96 | 2.93e-14 | 3.22e-13 | 13NEAT1, SLC11A1, LAPTM5, PLAUR, TYROBP, PSAP, CD14, ZEB2, ITGB2, CD68, SRGN, S100A4, FCER1G |
117 |
| TRAVAGLINI_LUNG_MACROPHAGE_CELL | 1.94e-20 | 38.41 | 19.91 | 4.35e-18 | 1.30e-17 | 18APOE, APOC1, CTSD, ACP5, CTSB, SLC11A1, IL1RN, GPNMB, CAPG, PLAUR, TYROBP, MMP19, PSAP, TGFBI, CSTB, RAB31, S100A10, FCER1G |
201 |
| TRAVAGLINI_LUNG_TREM2_DENDRITIC_CELL | 1.52e-25 | 29.28 | 16.04 | 1.02e-22 | 1.02e-22 | 28SPP1, APOE, APOC1, CTSD, ACP5, CTSB, CTSZ, NEAT1, SLC11A1, HMOX1, ITGAX, GPNMB, CAPG, PLAUR, TYROBP, PSAP, CD14, TFRC, TGFBI, CSTB, ITGB2, SLC16A3, RAB31, CD68, S100A10, FCER1G, RAB20, GNAS |
572 |
| AIZARANI_LIVER_C25_KUPFFER_CELLS_4 | 2.89e-15 | 30.36 | 14.83 | 1.38e-13 | 1.94e-12 | 14SLC11A1, ITGAX, IL1RN, LAPTM5, PLAUR, TYROBP, PSAP, RGS1, TGFBI, ZEB2, ITGB2, RAB31, SRGN, FCER1G |
174 |
| AIZARANI_LIVER_C23_KUPFFER_CELLS_3 | 1.10e-16 | 28.69 | 14.54 | 9.22e-15 | 7.37e-14 | 16CTSB, SLC2A3, SLC11A1, ITGAX, LAPTM5, PLAUR, TYROBP, PSAP, CD14, ZEB2, ITGB2, RAB31, S100A10, SRGN, S100A4, FCER1G |
221 |
| AIZARANI_LIVER_C6_KUPFFER_CELLS_2 | 1.79e-16 | 27.75 | 14.07 | 1.20e-14 | 1.20e-13 | 16CTSD, CTSB, SLC11A1, HMOX1, ITGAX, GPNMB, LAPTM5, TYROBP, PSAP, CD14, RGS1, TGFBI, ZEB2, ITGB2, RAB31, FCER1G |
228 |
| TRAVAGLINI_LUNG_EREG_DENDRITIC_CELL | 1.49e-22 | 24.52 | 13.43 | 4.99e-20 | 9.97e-20 | 26CTSB, CTSZ, NEAT1, SLC2A3, SLC11A1, HMOX1, ITGAX, IL1RN, SLC16A10, PLAUR, TYROBP, MMP19, CHST11, PSAP, CD14, TFRC, TGFBI, ZEB2, SLC16A3, RAB31, S100A10, SRGN, FCER1G, RAB20, HIF1A, ELL2 |
579 |
| CUI_DEVELOPING_HEART_C8_MACROPHAGE | 1.37e-16 | 24.98 | 12.86 | 1.02e-14 | 9.17e-14 | 17CTSD, CTSB, NEAT1, SLC2A3, ITGAX, LAPTM5, PLAUR, TYROBP, PSAP, CD14, RGS1, ITGB2, RAB31, CD68, SRGN, S100A4, FCER1G |
275 |
| AIZARANI_LIVER_C18_NK_NKT_CELLS_5 | 3.22e-11 | 28.12 | 12.23 | 1.03e-09 | 2.16e-08 | 10SLC11A1, ITGAX, LAPTM5, PLAUR, TYROBP, PSAP, TGFBI, ZEB2, RAB31, FCER1G |
121 |
| FAN_OVARY_CL13_MONOCYTE_MACROPHAGE | 2.31e-19 | 22.54 | 12.18 | 3.10e-17 | 1.55e-16 | 22CTSD, CTSB, CTSZ, NEAT1, HMOX1, CAPG, LAPTM5, PLAUR, TYROBP, PSAP, CD14, RGS1, ZEB2, CSTB, ITGB2, SLC16A3, CD68, S100A10, SRGN, S100A4, FCER1G, B2M |
458 |
| TRAVAGLINI_LUNG_OLR1_CLASSICAL_MONOCYTE_CELL | 3.47e-20 | 19.36 | 10.63 | 5.81e-18 | 2.33e-17 | 26CTSB, NEAT1, SLC2A3, SLC11A1, HMOX1, ITGAX, IL1RN, SLC16A10, PLAUR, TYROBP, MMP19, CHST11, PSAP, CD14, TFRC, TGFBI, ZEB2, CSTB, SLC16A3, RAB31, S100A10, SRGN, FCER1G, RAB20, HIF1A, ELL2 |
726 |
| FAN_EMBRYONIC_CTX_BIG_GROUPS_MICROGLIA | 9.14e-16 | 19.93 | 10.42 | 5.11e-14 | 6.13e-13 | 18SPP1, APOE, APOC1, CTSD, CTSB, NEAT1, HMOX1, ITGAX, LAPTM5, TYROBP, PSAP, CD14, RGS1, ITGB2, CD68, SRGN, FCER1G, B2M |
371 |
| AIZARANI_LIVER_C31_KUPFFER_CELLS_5 | 8.20e-10 | 24.70 | 10.30 | 2.04e-08 | 5.50e-07 | 9CTSB, SLC11A1, ITGAX, IL1RN, PLAUR, TYROBP, PSAP, ZEB2, FCER1G |
120 |
| HU_FETAL_RETINA_MICROGLIA | 1.50e-15 | 19.33 | 10.11 | 7.75e-14 | 1.01e-12 | 18SPP1, CTSD, CTSB, NEAT1, SLC2A3, HMOX1, ITGAX, LAPTM5, PLAUR, TYROBP, PSAP, CD14, RGS1, ITGB2, RAB31, SRGN, FCER1G, B2M |
382 |
| DESCARTES_FETAL_THYMUS_ANTIGEN_PRESENTING_CELLS | 4.74e-11 | 21.89 | 9.93 | 1.45e-09 | 3.18e-08 | 11ITGAX, IL1RN, PLAUR, TYROBP, CD14, RGS1, SLC16A3, RAB31, CD68, SRGN, FCER1G |
172 |
| MENON_FETAL_KIDNEY_10_IMMUNE_CELLS | 3.25e-08 | 26.73 | 9.84 | 7.04e-07 | 2.18e-05 | 7NEAT1, LAPTM5, TYROBP, PSAP, SRGN, S100A4, B2M |
83 |
| AIZARANI_LIVER_C2_KUPFFER_CELLS_1 | 1.15e-11 | 20.99 | 9.83 | 4.06e-10 | 7.72e-09 | 12CTSB, ITGAX, LAPTM5, PLAUR, TYROBP, PSAP, RGS1, TGFBI, ITGB2, RAB31, SRGN, FCER1G |
200 |
| MANNO_MIDBRAIN_NEUROTYPES_HMGL | 2.65e-17 | 17.71 | 9.59 | 2.54e-15 | 1.78e-14 | 22SPP1, APOE, APOC1, CTSD, CTSB, NEAT1, HMOX1, GPNMB, CAPG, LAPTM5, PLAUR, TYROBP, CD14, RGS1, ARHGAP26, ITGB2, CD68, SRGN, FCER1G, RREB1, ELL2, B2M |
577 |
| DESCARTES_FETAL_CEREBELLUM_MICROGLIA | 1.37e-17 | 17.23 | 9.37 | 1.53e-15 | 9.16e-15 | 23SPP1, APOE, APOC1, MITF, CTSB, CTSZ, SLC11A1, HMOX1, ITGAX, IL1RN, GPNMB, CAPG, LAPTM5, TYROBP, CD14, RGS1, ITGB2, CD68, S100A4, FCER1G, RAB20, ELL2, HK2 |
642 |
Dowload full table
mSigDB Hallmark Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| HALLMARK_HYPOXIA | 1.19e-05 | 10.54 | 3.95 | 5.94e-04 | 5.94e-04 | 7SLC2A3, HMOX1, PLAUR, TGFBI, PAM, S100A4, HK2 |
200 |
| HALLMARK_COMPLEMENT | 1.24e-04 | 8.78 | 3.02 | 2.06e-03 | 6.18e-03 | 6MMP12, APOC1, CTSD, CTSB, PLAUR, FCER1G |
200 |
| HALLMARK_KRAS_SIGNALING_UP | 1.24e-04 | 8.78 | 3.02 | 2.06e-03 | 6.18e-03 | 6SPP1, GPNMB, LAPTM5, PLAUR, ITGB2, FCER1G |
200 |
| HALLMARK_IL2_STAT5_SIGNALING | 1.06e-03 | 7.16 | 2.19 | 1.08e-02 | 5.28e-02 | 5SPP1, CTSZ, SLC2A3, CAPG, HK2 |
199 |
| HALLMARK_ALLOGRAFT_REJECTION | 1.08e-03 | 7.12 | 2.18 | 1.08e-02 | 5.40e-02 | 5CAPG, ITGB2, SRGN, HIF1A, B2M |
200 |
| HALLMARK_ANGIOGENESIS | 8.94e-03 | 15.30 | 1.73 | 3.73e-02 | 4.47e-01 | 2SPP1, S100A4 |
36 |
| HALLMARK_MTORC1_SIGNALING | 7.71e-03 | 5.55 | 1.44 | 3.50e-02 | 3.85e-01 | 4SLC2A3, TFRC, ITGB2, HK2 |
200 |
| HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | 7.71e-03 | 5.55 | 1.44 | 3.50e-02 | 3.85e-01 | 4SPP1, CAPG, PLAUR, TGFBI |
200 |
| HALLMARK_INFLAMMATORY_RESPONSE | 7.71e-03 | 5.55 | 1.44 | 3.50e-02 | 3.85e-01 | 4PLAUR, CD14, RGS1, HIF1A |
200 |
| HALLMARK_GLYCOLYSIS | 7.71e-03 | 5.55 | 1.44 | 3.50e-02 | 3.85e-01 | 4TGFBI, SLC16A3, PAM, HK2 |
200 |
| HALLMARK_P53_PATHWAY | 7.71e-03 | 5.55 | 1.44 | 3.50e-02 | 3.85e-01 | 4CTSD, HMOX1, S100A10, S100A4 |
200 |
| HALLMARK_HEME_METABOLISM | 7.71e-03 | 5.55 | 1.44 | 3.50e-02 | 3.85e-01 | 4ACP5, CTSB, TFRC, ELL2 |
200 |
| HALLMARK_IL6_JAK_STAT3_SIGNALING | 4.64e-02 | 6.13 | 0.71 | 1.78e-01 | 1.00e+00 | 2HMOX1, CD14 |
87 |
| HALLMARK_PROTEIN_SECRETION | 5.52e-02 | 5.54 | 0.64 | 1.97e-01 | 1.00e+00 | 2PAM, GNAS |
96 |
| HALLMARK_ANDROGEN_RESPONSE | 5.93e-02 | 5.31 | 0.62 | 1.98e-01 | 1.00e+00 | 2ELL2, B2M |
100 |
| HALLMARK_COAGULATION | 1.03e-01 | 3.83 | 0.45 | 3.21e-01 | 1.00e+00 | 2APOC1, CTSB |
138 |
| HALLMARK_UV_RESPONSE_UP | 1.28e-01 | 3.34 | 0.39 | 3.67e-01 | 1.00e+00 | 2HMOX1, TFRC |
158 |
| HALLMARK_APOPTOSIS | 1.32e-01 | 3.28 | 0.38 | 3.67e-01 | 1.00e+00 | 2HMOX1, CD14 |
161 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 1.85e-01 | 2.63 | 0.31 | 4.02e-01 | 1.00e+00 | 2SLC2A3, PLAUR |
200 |
| HALLMARK_ADIPOGENESIS | 1.85e-01 | 2.63 | 0.31 | 4.02e-01 | 1.00e+00 | 2APOE, RREB1 |
200 |
Dowload full table
KEGG Pathways:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| KEGG_LYSOSOME | 2.10e-08 | 21.07 | 8.35 | 3.91e-06 | 3.91e-06 | 8CTSD, ACP5, CTSB, CTSZ, SLC11A1, LAPTM5, PSAP, CD68 |
121 |
| KEGG_NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY | 1.67e-02 | 5.95 | 1.17 | 1.00e+00 | 1.00e+00 | 3TYROBP, ITGB2, FCER1G |
137 |
| KEGG_REGULATION_OF_ACTIN_CYTOSKELETON | 5.07e-02 | 3.80 | 0.75 | 1.00e+00 | 1.00e+00 | 3ITGAX, CD14, ITGB2 |
213 |
| KEGG_HEMATOPOIETIC_CELL_LINEAGE | 4.64e-02 | 6.13 | 0.71 | 1.00e+00 | 1.00e+00 | 2CD14, TFRC |
87 |
| KEGG_ANTIGEN_PROCESSING_AND_PRESENTATION | 4.73e-02 | 6.05 | 0.70 | 1.00e+00 | 1.00e+00 | 2CTSB, B2M |
88 |
| KEGG_MELANOGENESIS | 6.04e-02 | 5.26 | 0.61 | 1.00e+00 | 1.00e+00 | 2MITF, GNAS |
101 |
| KEGG_TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY | 6.14e-02 | 5.21 | 0.60 | 1.00e+00 | 1.00e+00 | 2SPP1, CD14 |
102 |
| KEGG_SULFUR_METABOLISM | 5.05e-02 | 21.22 | 0.49 | 1.00e+00 | 1.00e+00 | 1CHST11 |
13 |
| KEGG_RIBOFLAVIN_METABOLISM | 6.18e-02 | 16.98 | 0.40 | 1.00e+00 | 1.00e+00 | 1ACP5 |
16 |
| KEGG_ENDOCYTOSIS | 1.59e-01 | 2.91 | 0.34 | 1.00e+00 | 1.00e+00 | 2TFRC, RAB31 |
181 |
| KEGG_GLYCOSAMINOGLYCAN_BIOSYNTHESIS_CHONDROITIN_SULFATE | 8.40e-02 | 12.14 | 0.29 | 1.00e+00 | 1.00e+00 | 1CHST11 |
22 |
| KEGG_GALACTOSE_METABOLISM | 9.85e-02 | 10.19 | 0.24 | 1.00e+00 | 1.00e+00 | 1HK2 |
26 |
| KEGG_ASTHMA | 1.13e-01 | 8.79 | 0.21 | 1.00e+00 | 1.00e+00 | 1FCER1G |
30 |
| KEGG_PATHWAYS_IN_CANCER | 3.61e-01 | 1.61 | 0.19 | 1.00e+00 | 1.00e+00 | 2MITF, HIF1A |
325 |
| KEGG_FRUCTOSE_AND_MANNOSE_METABOLISM | 1.27e-01 | 7.73 | 0.19 | 1.00e+00 | 1.00e+00 | 1HK2 |
34 |
| KEGG_PORPHYRIN_AND_CHLOROPHYLL_METABOLISM | 1.51e-01 | 6.37 | 0.15 | 1.00e+00 | 1.00e+00 | 1HMOX1 |
41 |
| KEGG_AMINO_SUGAR_AND_NUCLEOTIDE_SUGAR_METABOLISM | 1.61e-01 | 5.93 | 0.14 | 1.00e+00 | 1.00e+00 | 1HK2 |
44 |
| KEGG_VASOPRESSIN_REGULATED_WATER_REABSORPTION | 1.61e-01 | 5.93 | 0.14 | 1.00e+00 | 1.00e+00 | 1GNAS |
44 |
| KEGG_TYPE_II_DIABETES_MELLITUS | 1.71e-01 | 5.54 | 0.13 | 1.00e+00 | 1.00e+00 | 1HK2 |
47 |
| KEGG_TASTE_TRANSDUCTION | 1.87e-01 | 5.00 | 0.12 | 1.00e+00 | 1.00e+00 | 1GNAS |
52 |
Dowload full table
CHR Positional Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| chr5q31 | 1.01e-01 | 2.81 | 0.56 | 1.00e+00 | 1.00e+00 | 3CD14, ARHGAP26, TGFBI |
287 |
| chr3q29 | 8.36e-02 | 4.34 | 0.51 | 1.00e+00 | 1.00e+00 | 2TM4SF19, TFRC |
122 |
| chr10q22 | 1.88e-01 | 2.60 | 0.30 | 1.00e+00 | 1.00e+00 | 2PSAP, SRGN |
202 |
| chr19q13 | 1.00e+00 | 0.94 | 0.24 | 1.00e+00 | 1.00e+00 | 4APOE, APOC1, PLAUR, TYROBP |
1165 |
| chr21q22 | 3.99e-01 | 1.48 | 0.17 | 1.00e+00 | 1.00e+00 | 2CSTB, ITGB2 |
353 |
| chr3p13 | 1.47e-01 | 6.54 | 0.16 | 1.00e+00 | 1.00e+00 | 1MITF |
40 |
| chr1q21 | 6.64e-01 | 1.34 | 0.16 | 1.00e+00 | 1.00e+00 | 2S100A10, S100A4 |
392 |
| chr20q13 | 6.68e-01 | 1.31 | 0.15 | 1.00e+00 | 1.00e+00 | 2CTSZ, GNAS |
400 |
| chr2p12 | 1.90e-01 | 4.91 | 0.12 | 1.00e+00 | 1.00e+00 | 1HK2 |
53 |
| chr6q13 | 2.00e-01 | 4.64 | 0.11 | 1.00e+00 | 1.00e+00 | 1CD109 |
56 |
| chr6p24 | 2.03e-01 | 4.55 | 0.11 | 1.00e+00 | 1.00e+00 | 1RREB1 |
57 |
| chr5q15 | 2.13e-01 | 4.32 | 0.11 | 1.00e+00 | 1.00e+00 | 1ELL2 |
60 |
| chr5q21 | 2.25e-01 | 4.05 | 0.10 | 1.00e+00 | 1.00e+00 | 1PAM |
64 |
| chr2q22 | 2.37e-01 | 3.81 | 0.09 | 1.00e+00 | 1.00e+00 | 1ZEB2 |
68 |
| chr4q22 | 2.43e-01 | 3.70 | 0.09 | 1.00e+00 | 1.00e+00 | 1SPP1 |
70 |
| chr1q31 | 2.46e-01 | 3.64 | 0.09 | 1.00e+00 | 1.00e+00 | 1RGS1 |
71 |
| chr7p15 | 3.17e-01 | 2.68 | 0.07 | 1.00e+00 | 1.00e+00 | 1GPNMB |
96 |
| chr11q22 | 3.23e-01 | 2.63 | 0.06 | 1.00e+00 | 1.00e+00 | 1MMP12 |
98 |
| chr6q21 | 3.72e-01 | 2.20 | 0.05 | 1.00e+00 | 1.00e+00 | 1SLC16A10 |
117 |
| chr14q23 | 3.89e-01 | 2.07 | 0.05 | 1.00e+00 | 1.00e+00 | 1HIF1A |
124 |
Dowload full table
Transcription Factor Targets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| TCANNTGAY_SREBP1_01 | 7.44e-05 | 5.84 | 2.48 | 8.43e-02 | 8.43e-02 | 9CTSD, HMOX1, GPNMB, CD109, TM4SF19, PSAP, RGS1, HIF1A, HK2 |
479 |
| HES2_TARGET_GENES | 1.14e-03 | 3.12 | 1.52 | 5.01e-01 | 1.00e+00 | 13CTSD, MITF, CTSB, NEAT1, HMOX1, CD109, MMP19, TFRC, ARHGAP26, TGFBI, RAB31, S100A10, SRGN |
1420 |
| TFEB_TARGET_GENES | 3.04e-03 | 2.87 | 1.36 | 7.60e-01 | 1.00e+00 | 12CTSD, ACP5, CTSB, NEAT1, SLC2A3, HMOX1, ITGAX, TGFBI, RAB31, CD68, PAM, HIF1A |
1387 |
| SMN1_SMN2_TARGET_GENES | 6.24e-03 | 3.01 | 1.28 | 9.24e-01 | 1.00e+00 | 9CTSB, CAPG, TFRC, ARHGAP26, ITGB2, RAB31, S100A4, GNAS, B2M |
922 |
| IRF2_01 | 1.42e-02 | 6.33 | 1.24 | 9.24e-01 | 1.00e+00 | 3MITF, TRPM3, B2M |
129 |
| TGANTCA_AP1_C | 7.29e-03 | 2.77 | 1.23 | 9.24e-01 | 1.00e+00 | 10MMP12, MITF, SLC11A1, ITGAX, IL1RN, GPNMB, TM4SF19, MMP19, CD68, S100A10 |
1139 |
| USF_Q6_01 | 1.50e-02 | 4.53 | 1.17 | 9.24e-01 | 1.00e+00 | 4HMOX1, RAB31, GNAS, HIF1A |
244 |
| GCM_Q2 | 1.52e-02 | 4.51 | 1.17 | 9.24e-01 | 1.00e+00 | 4MITF, CHST11, GNAS, ELL2 |
245 |
| SNRNP70_TARGET_GENES | 1.06e-02 | 2.75 | 1.17 | 9.24e-01 | 1.00e+00 | 9APOE, NEAT1, TFRC, ARHGAP26, ZEB2, SLC16A3, RAB31, CD68, B2M |
1009 |
| CEBPDELTA_Q6 | 1.56e-02 | 4.47 | 1.16 | 9.24e-01 | 1.00e+00 | 4MITF, IL1RN, TRPM3, ZEB2 |
247 |
| IRF1_01 | 1.73e-02 | 4.33 | 1.12 | 9.24e-01 | 1.00e+00 | 4MITF, TRPM3, CHST11, B2M |
255 |
| CEBPA_01 | 1.75e-02 | 4.31 | 1.12 | 9.24e-01 | 1.00e+00 | 4MITF, CD109, FCER1G, B2M |
256 |
| TGGAAA_NFAT_Q4_01 | 1.89e-02 | 2.29 | 1.11 | 9.24e-01 | 1.00e+00 | 13SPP1, MITF, IL1RN, TRPM3, CHST11, RGS1, ZEB2, CD68, S100A10, FCER1G, GNAS, HIF1A, RREB1 |
1934 |
| USF_01 | 1.93e-02 | 4.18 | 1.09 | 9.24e-01 | 1.00e+00 | 4HMOX1, TFRC, RAB31, GNAS |
264 |
| AP1_Q2 | 1.96e-02 | 4.16 | 1.08 | 9.24e-01 | 1.00e+00 | 4IL1RN, LAPTM5, MMP19, S100A10 |
265 |
| AP1_Q6_01 | 2.00e-02 | 4.13 | 1.07 | 9.24e-01 | 1.00e+00 | 4SLC11A1, IL1RN, LAPTM5, MMP19 |
267 |
| AP1FJ_Q2 | 2.10e-02 | 4.07 | 1.06 | 9.24e-01 | 1.00e+00 | 4IL1RN, LAPTM5, MMP19, S100A10 |
271 |
| AP1_Q4 | 2.10e-02 | 4.07 | 1.06 | 9.24e-01 | 1.00e+00 | 4IL1RN, LAPTM5, MMP19, S100A10 |
271 |
| GR_Q6 | 2.20e-02 | 4.01 | 1.04 | 9.24e-01 | 1.00e+00 | 4MITF, CD109, ZEB2, SRGN |
275 |
| GREB1_TARGET_GENES | 2.14e-02 | 2.55 | 1.03 | 9.24e-01 | 1.00e+00 | 8CTSB, GPNMB, TGFBI, ITGB2, SLC16A3, S100A10, PAM, S100A4 |
941 |
Dowload full table
GO Biological Processes:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GOBP_POSITIVE_REGULATION_OF_RECEPTOR_CATABOLIC_PROCESS | 3.22e-04 | 103.49 | 9.64 | 7.53e-02 | 1.00e+00 | 2APOE, LAPTM5 |
7 |
| GOBP_CELLULAR_RESPONSE_TO_LIPOPROTEIN_PARTICLE_STIMULUS | 9.37e-06 | 36.12 | 8.90 | 7.01e-03 | 7.01e-02 | 4APOE, ITGB2, CD68, FCER1G |
34 |
| GOBP_WOUND_HEALING_INVOLVED_IN_INFLAMMATORY_RESPONSE | 4.28e-04 | 86.37 | 8.33 | 8.72e-02 | 1.00e+00 | 2HMOX1, HIF1A |
8 |
| GOBP_TRIGLYCERIDE_RICH_LIPOPROTEIN_PARTICLE_CLEARANCE | 4.28e-04 | 86.37 | 8.33 | 8.72e-02 | 1.00e+00 | 2APOE, APOC1 |
8 |
| GOBP_POSITIVE_REGULATION_OF_CHOLESTEROL_ESTERIFICATION | 5.49e-04 | 74.10 | 7.33 | 9.13e-02 | 1.00e+00 | 2APOE, APOC1 |
9 |
| GOBP_ELASTIN_METABOLIC_PROCESS | 5.49e-04 | 74.10 | 7.33 | 9.13e-02 | 1.00e+00 | 2MMP12, HIF1A |
9 |
| GOBP_CELLULAR_RESPONSE_TO_IRON_ION | 5.49e-04 | 74.10 | 7.33 | 9.13e-02 | 1.00e+00 | 2HMOX1, B2M |
9 |
| GOBP_VERY_LOW_DENSITY_LIPOPROTEIN_PARTICLE_CLEARANCE | 6.85e-04 | 64.86 | 6.55 | 1.05e-01 | 1.00e+00 | 2APOE, APOC1 |
10 |
| GOBP_COLLAGEN_CATABOLIC_PROCESS | 3.46e-05 | 25.26 | 6.32 | 1.62e-02 | 2.58e-01 | 4MMP12, CTSD, CTSB, MMP19 |
47 |
| GOBP_MYELOID_LEUKOCYTE_MEDIATED_IMMUNITY | 8.33e-12 | 12.09 | 6.27 | 3.12e-08 | 6.23e-08 | 17CTSD, CTSB, CTSZ, SLC2A3, SLC11A1, HMOX1, ITGAX, PLAUR, TYROBP, PSAP, CD14, CSTB, ITGB2, RAB31, CD68, FCER1G, B2M |
550 |
| GOBP_RESPONSE_TO_IRON_ION | 1.81e-04 | 31.85 | 5.95 | 5.27e-02 | 1.00e+00 | 3HMOX1, HIF1A, B2M |
28 |
| GOBP_REGULATION_OF_DENDRITIC_CELL_ANTIGEN_PROCESSING_AND_PRESENTATION | 8.35e-04 | 57.69 | 5.91 | 1.18e-01 | 1.00e+00 | 2SLC11A1, CD68 |
11 |
| GOBP_REGULATION_OF_TRANSCRIPTION_FROM_RNA_POLYMERASE_II_PROMOTER_IN_RESPONSE_TO_OXIDATIVE_STRESS | 8.35e-04 | 57.69 | 5.91 | 1.18e-01 | 1.00e+00 | 2HMOX1, HIF1A |
11 |
| GOBP_REGULATION_OF_RECEPTOR_CATABOLIC_PROCESS | 8.35e-04 | 57.69 | 5.91 | 1.18e-01 | 1.00e+00 | 2APOE, LAPTM5 |
11 |
| GOBP_SECRETION | 2.58e-14 | 10.23 | 5.63 | 1.93e-10 | 1.93e-10 | 27SPP1, APOE, CTSD, CTSB, CTSZ, SLC2A3, SLC11A1, HMOX1, ITGAX, IL1RN, SLC16A10, PLAUR, CD109, TYROBP, PSAP, CD14, CSTB, ITGB2, RAB31, CD68, SRGN, PAM, FCER1G, GNAS, HIF1A, B2M, HK2 |
1464 |
| GOBP_NEGATIVE_REGULATION_OF_LONG_TERM_SYNAPTIC_POTENTIATION | 9.99e-04 | 51.89 | 5.39 | 1.31e-01 | 1.00e+00 | 2APOE, TYROBP |
12 |
| GOBP_CELL_ACTIVATION_INVOLVED_IN_IMMUNE_RESPONSE | 4.94e-11 | 10.00 | 5.26 | 1.23e-07 | 3.69e-07 | 18CTSD, CTSB, CTSZ, SLC2A3, SLC11A1, HMOX1, ITGAX, PLAUR, TYROBP, PSAP, CD14, TFRC, CSTB, ITGB2, RAB31, CD68, FCER1G, B2M |
722 |
| GOBP_MYELOID_LEUKOCYTE_ACTIVATION | 1.26e-10 | 10.04 | 5.21 | 2.14e-07 | 9.44e-07 | 17CTSD, CTSB, CTSZ, SLC2A3, SLC11A1, HMOX1, ITGAX, PLAUR, TYROBP, PSAP, CD14, CSTB, ITGB2, RAB31, CD68, FCER1G, B2M |
659 |
| GOBP_IRON_ION_HOMEOSTASIS | 2.16e-05 | 17.14 | 5.17 | 1.35e-02 | 1.62e-01 | 5SLC11A1, HMOX1, TFRC, HIF1A, B2M |
86 |
| GOBP_PHOSPHOLIPID_EFFLUX | 1.18e-03 | 47.19 | 4.96 | 1.36e-01 | 1.00e+00 | 2APOE, APOC1 |
13 |
Dowload full table
Immunological Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GSE29618_MONOCYTE_VS_PDC_UP | 1.85e-14 | 26.27 | 12.86 | 4.82e-11 | 8.99e-11 | 14NEAT1, SLC2A3, HMOX1, ITGAX, IL1RN, TYROBP, CD14, ITGB2, RAB31, S100A10, S100A4, RAB20, HIF1A, HK2 |
199 |
| GSE10325_LUPUS_BCELL_VS_LUPUS_MYELOID_DN | 1.98e-14 | 26.13 | 12.79 | 4.82e-11 | 9.64e-11 | 14CTSD, CTSB, SLC11A1, HMOX1, IL1RN, PLAUR, TYROBP, PSAP, ARHGAP26, TGFBI, RAB31, S100A4, FCER1G, HK2 |
200 |
| GSE10325_LUPUS_CD4_TCELL_VS_LUPUS_MYELOID_DN | 5.03e-13 | 23.47 | 11.26 | 4.90e-10 | 2.45e-09 | 13CTSD, CTSB, SLC11A1, HMOX1, ITGAX, IL1RN, PLAUR, TYROBP, PSAP, CD14, TGFBI, RAB31, FCER1G |
200 |
| GSE22886_NAIVE_BCELL_VS_MONOCYTE_DN | 5.03e-13 | 23.47 | 11.26 | 4.90e-10 | 2.45e-09 | 13CTSD, CTSB, SLC11A1, CAPG, TYROBP, PSAP, CD14, SLC16A3, RAB31, S100A10, SRGN, S100A4, FCER1G |
200 |
| GSE22886_NAIVE_CD8_TCELL_VS_MONOCYTE_DN | 5.03e-13 | 23.47 | 11.26 | 4.90e-10 | 2.45e-09 | 13CTSB, SLC11A1, HMOX1, CAPG, LAPTM5, TYROBP, PSAP, CD14, ARHGAP26, TGFBI, SLC16A3, RAB31, FCER1G |
200 |
| GSE24634_TREG_VS_TCONV_POST_DAY10_IL4_CONVERSION_DN | 1.15e-11 | 20.99 | 9.83 | 8.00e-09 | 5.60e-08 | 12SPP1, APOC1, CTSB, CTSZ, HMOX1, ITGAX, CAPG, TYROBP, PSAP, CD14, ITGB2, RAB31 |
200 |
| GSE29618_MONOCYTE_VS_MDC_UP | 1.15e-11 | 20.99 | 9.83 | 8.00e-09 | 5.60e-08 | 12CTSD, CTSB, SLC11A1, HMOX1, TYROBP, PSAP, CD14, CD68, S100A4, FCER1G, HIF1A, ELL2 |
200 |
| GSE24634_TEFF_VS_TCONV_DAY10_IN_CULTURE_DN | 2.35e-10 | 18.65 | 8.48 | 1.43e-07 | 1.15e-06 | 11SPP1, CTSZ, SLC11A1, ITGAX, GPNMB, CAPG, PSAP, ZEB2, ITGB2, RAB31, FCER1G |
200 |
| GSE34156_TLR1_TLR2_LIGAND_VS_NOD2_AND_TLR1_TLR2_LIGAND_24H_TREATED_MONOCYTE_UP | 3.72e-09 | 16.71 | 7.34 | 1.74e-06 | 1.81e-05 | 10CTSD, CTSZ, SLC11A1, HMOX1, CAPG, TYROBP, PSAP, CD14, TGFBI, RAB31 |
197 |
| GSE24634_TREG_VS_TCONV_POST_DAY7_IL4_CONVERSION_DN | 3.90e-09 | 16.62 | 7.30 | 1.74e-06 | 1.90e-05 | 10MMP12, ACP5, CTSB, CTSZ, PLAUR, TYROBP, PSAP, CD14, CD68, RAB20 |
198 |
| GSE6269_FLU_VS_STAPH_AUREUS_INF_PBMC_DN | 1.48e-08 | 17.36 | 7.29 | 5.54e-06 | 7.21e-05 | 9CTSD, CTSB, SLC11A1, IL1RN, PLAUR, S100A10, SRGN, RAB20, HIF1A |
167 |
| GSE29618_BCELL_VS_MDC_DN | 4.29e-09 | 16.45 | 7.22 | 1.74e-06 | 2.09e-05 | 10CTSB, ITGAX, CAPG, RGS1, TGFBI, CSTB, ITGB2, RAB31, S100A10, S100A4 |
200 |
| GSE29618_PDC_VS_MDC_DN | 4.29e-09 | 16.45 | 7.22 | 1.74e-06 | 2.09e-05 | 10SLC2A3, ITGAX, IL1RN, CSTB, ITGB2, RAB31, S100A10, S100A4, HIF1A, HK2 |
200 |
| GSE9988_ANTI_TREM1_VS_CTRL_TREATED_MONOCYTES_UP | 6.34e-08 | 14.52 | 6.11 | 1.53e-05 | 3.09e-04 | 9SPP1, SLC2A3, PLAUR, MMP19, RGS1, TFRC, RAB20, RREB1, ELL2 |
198 |
| GSE10325_BCELL_VS_MYELOID_DN | 6.91e-08 | 14.37 | 6.05 | 1.53e-05 | 3.36e-04 | 9CTSB, SLC11A1, HMOX1, IL1RN, PLAUR, PSAP, TGFBI, SRGN, S100A4 |
200 |
| GSE17721_ALL_VS_24H_PAM3CSK4_BMDC_UP | 6.91e-08 | 14.37 | 6.05 | 1.53e-05 | 3.36e-04 | 9SPP1, APOE, MITF, CTSB, ITGAX, IL1RN, CAPG, PLAUR, PSAP |
200 |
| GSE29618_BCELL_VS_MONOCYTE_DN | 6.91e-08 | 14.37 | 6.05 | 1.53e-05 | 3.36e-04 | 9CTSB, HMOX1, PLAUR, PSAP, CD14, ITGB2, RAB31, CD68, RAB20 |
200 |
| GSE29618_BCELL_VS_MONOCYTE_DAY7_FLU_VACCINE_DN | 6.91e-08 | 14.37 | 6.05 | 1.53e-05 | 3.36e-04 | 9CTSD, HMOX1, IL1RN, PLAUR, TYROBP, PSAP, CD14, CD68, RAB20 |
200 |
| GSE29618_BCELL_VS_MDC_DAY7_FLU_VACCINE_DN | 6.91e-08 | 14.37 | 6.05 | 1.53e-05 | 3.36e-04 | 9ITGAX, IL1RN, TYROBP, PSAP, TGFBI, RAB31, S100A10, S100A4, FCER1G |
200 |
| GSE3337_CTRL_VS_16H_IFNG_IN_CD8POS_DC_UP | 6.91e-08 | 14.37 | 6.05 | 1.53e-05 | 3.36e-04 | 9MMP12, APOE, CTSD, ACP5, CTSB, HMOX1, TYROBP, CD14, S100A4 |
200 |
Top Ranked Transcription Factors for this Gene Expression Program:
| Gene Symbol | TF Rank | DNA Binding Domain | Motif Status | IUPAC PWM | GTEx | DepMap | Decartes |
|---|---|---|---|---|---|---|---|
| MITF | 7 | Yes | Known motif | Monomer or homomultimer | 100 perc ID - in vitro | None | Binds DNA both as homo- and heterodimers (PMID: 23207919). The structure 4ATI is a homodimer with DNA GTTAGCACATGACCCT. |
| HMOX1 | 13 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Well characterized enzyme with no additional domains. Included only because GO lists it as a regulator of TFs. No support for DNA binding activity. |
| ZEB2 | 33 | Yes | Inferred motif | Monomer or homomultimer | In vivo/Misc source | None | None |
| ITGB2 | 35 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| HIF1A | 46 | Yes | Known motif | Obligate heteromer | In vivo/Misc source | Only known motifs are from Transfac or HocoMoco - origin is uncertain | Binds as obligate heteromer with ARNT (PMID: 9027737). |
| RREB1 | 47 | Yes | Known motif | Monomer or homomultimer | In vivo/Misc source | None | None |
| SGK1 | 58 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| RGCC | 64 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| MEIS1 | 67 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| CEBPB | 71 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| PLXNA4 | 95 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Protein is a semaphorin receptor that operates far upstream on the signaling cascade (PMID: 12591607). |
| CAMK2A | 99 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| ZBTB20 | 131 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| SPI1 | 132 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| TSHZ2 | 134 | Yes | Likely to be sequence specific TF | Monomer or homomultimer | No motif | None | Also contains an atypical homodomain (Uniprot) not identified by Pfam. |
| SORBS2 | 139 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| ZFP36L1 | 140 | No | ssDNA/RNA binding | Not a DNA binding protein | No motif | None | RNA-binding protein (PMID: 27102483; PMID: 17013884). |
| AFF1 | 157 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | Has a putative AT-hook | MLL fusion partner |
| LPP | 159 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | znfLIM is probably not a DNA binding domain (PMID: 8506279). The protein has been reported to be coactivator for ETV4 (PMID: 16738319). |
| ATF3 | 185 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
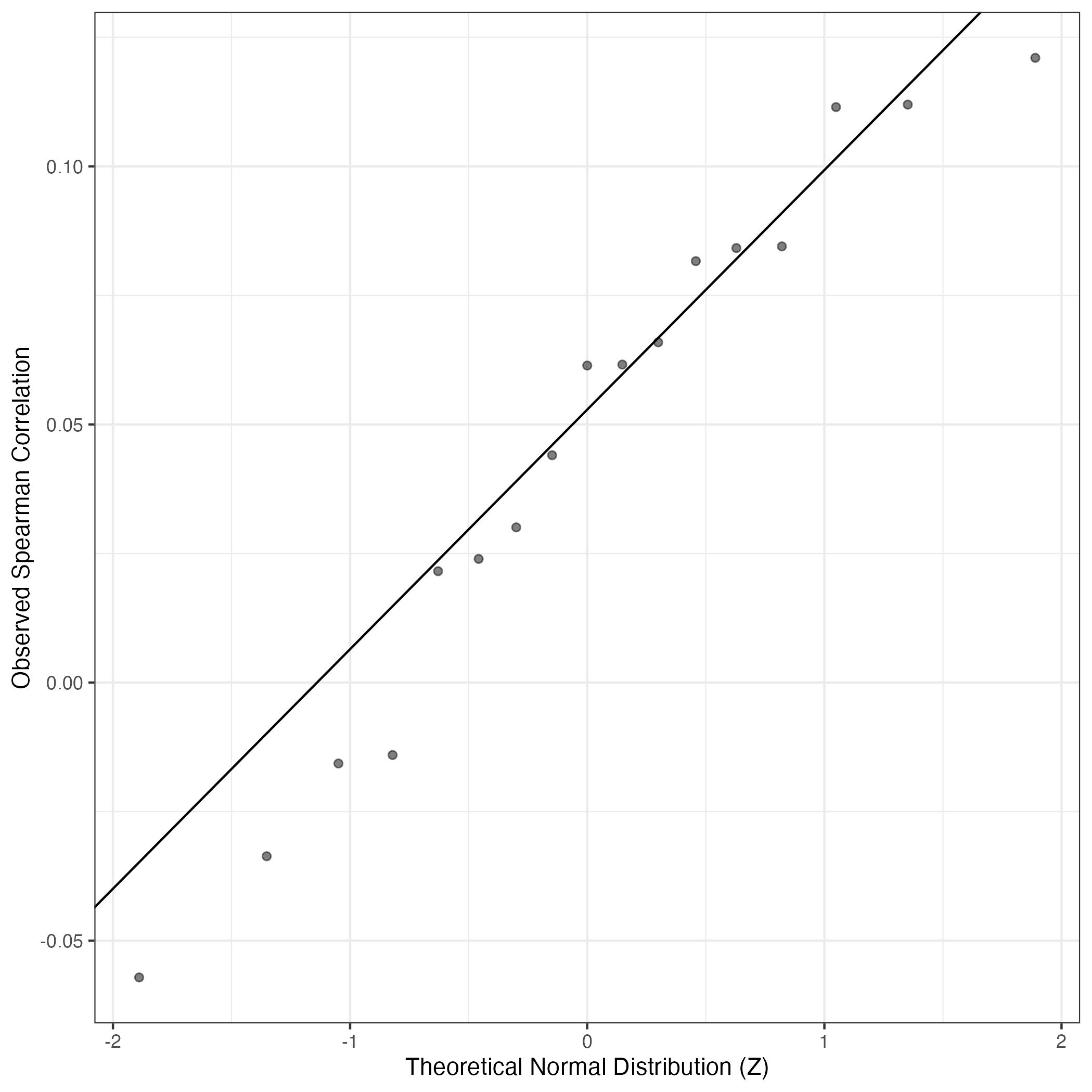
QQ Plot showing correlations with other Gene Expression Programs in this dataset, calculated by Spearman correlation:
Interactive QQ-plot of gene loadings:
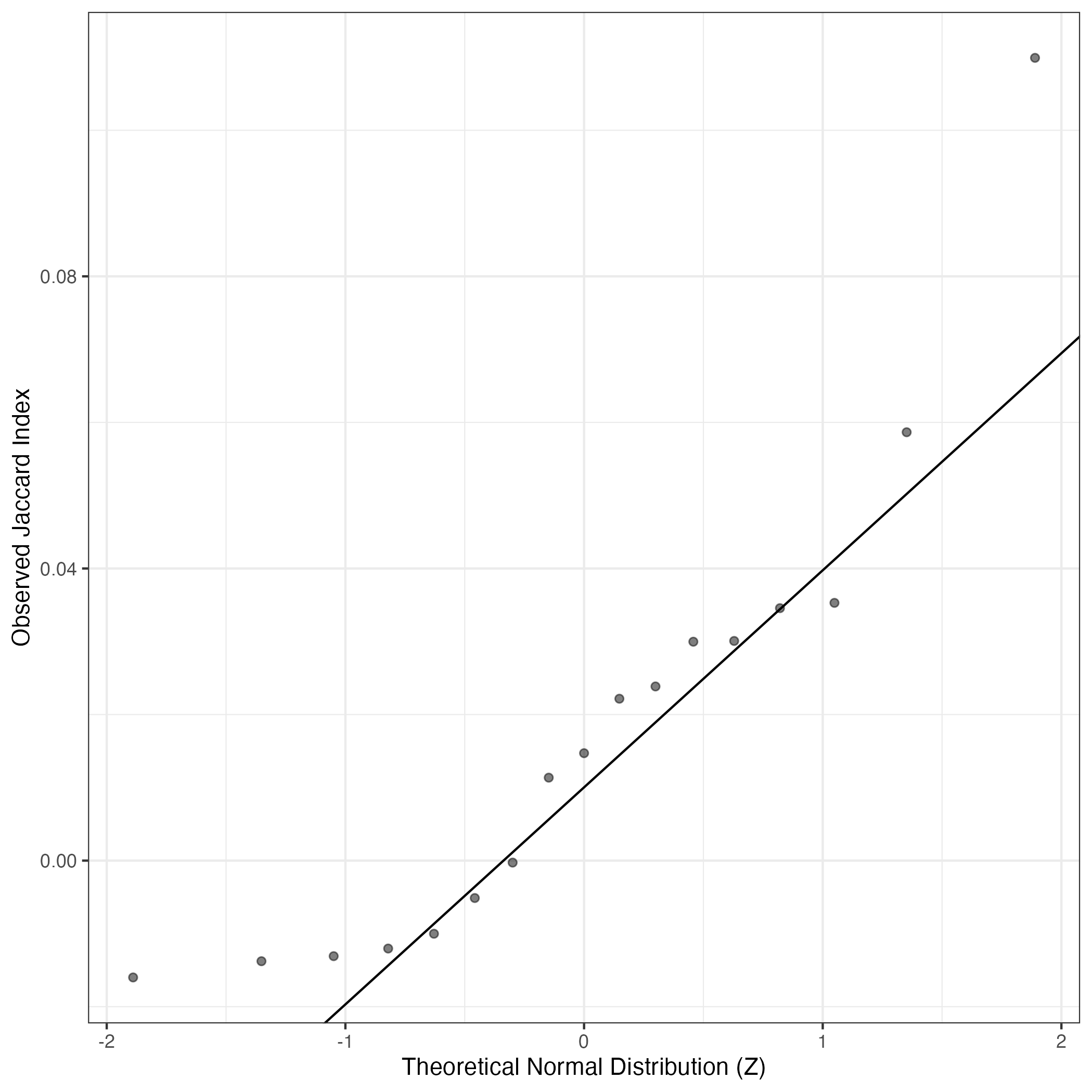
A similar QQ-plot as above, but only for instances where the H value is e.g. > 25, i.e. we are confident that the expression program is active above noise. Agreemenet between these binary vectors is tested using the Jaccard Index, with the P-values calculated by an exact test:
Interactive QQ-plot:
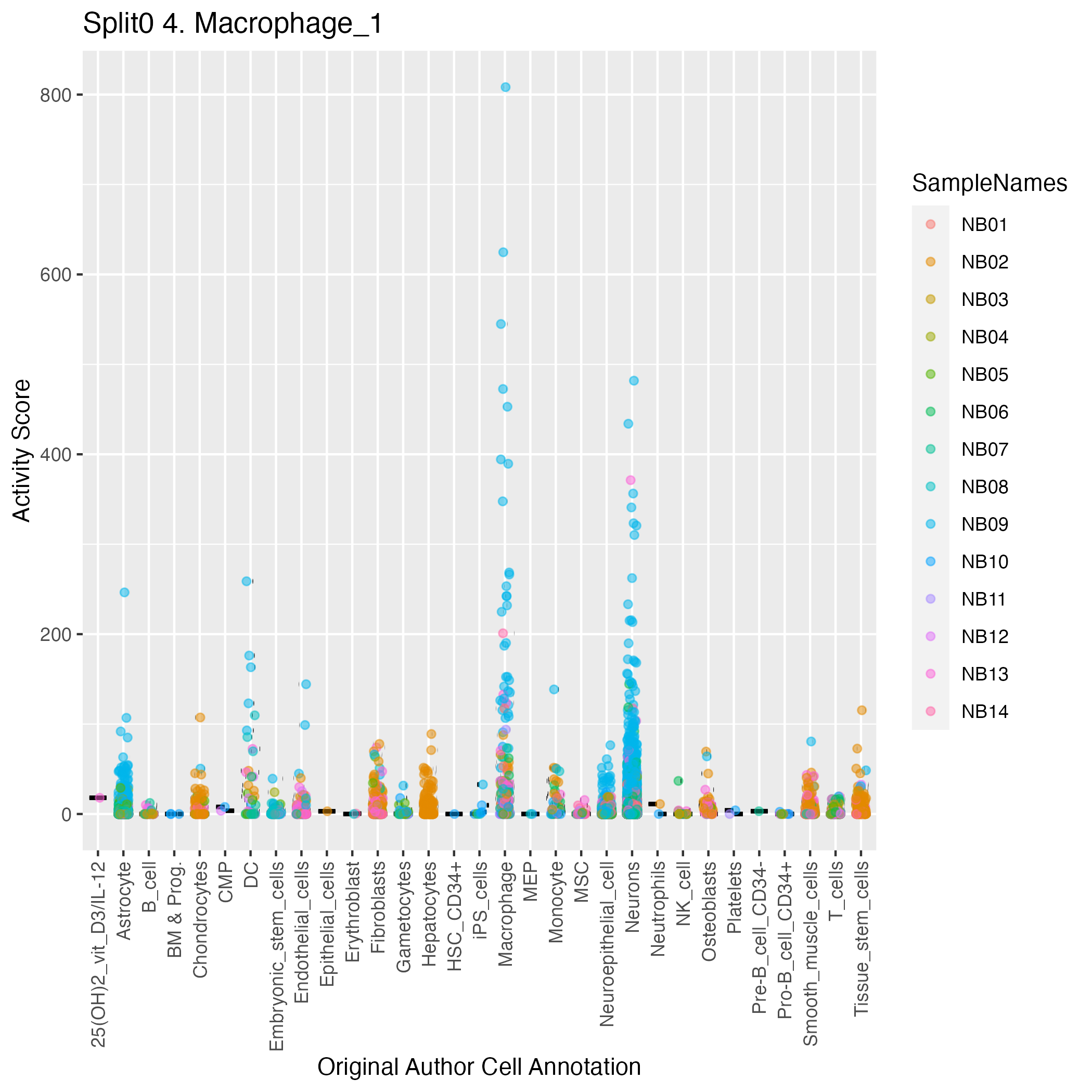
Singler cell type annotations for the top 50 cells on this program.
| Cell ID | Singler label | Singler Delta | Activity Score | Top Singler Raw Scores |
|---|---|---|---|---|
| NB09_ACTGATGGTTGGTAAA-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.11 | 808.25 | Raw ScoresMacrophage:monocyte-derived: 0.29, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.28, Macrophage:monocyte-derived:M-CSF: 0.28, Macrophage:monocyte-derived:IL-4/cntrl: 0.28, Macrophage:monocyte-derived:IL-4/TGFb: 0.28, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.28, Macrophage:monocyte-derived:S._aureus: 0.28, Monocyte:CXCL4: 0.28, DC:monocyte-derived:AM580: 0.28, Macrophage:monocyte-derived:IFNa: 0.28 |
| NB09_GTGAAGGAGTACGTTC-1 | Neurons:adrenal_medulla_cell_line | 0.06 | 624.65 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.2, Macrophage:monocyte-derived: 0.2, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.2, Monocyte:CXCL4: 0.2, Macrophage:monocyte-derived:IL-4/cntrl: 0.2, Macrophage:monocyte-derived:IL-4/TGFb: 0.2, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.19, Monocyte:MCSF: 0.19, Macrophage:monocyte-derived:M-CSF: 0.19, Macrophage:monocyte-derived:IFNa: 0.19 |
| NB09_TCTATTGAGATGAGAG-1 | Macrophage:Alveolar | 0.04 | 544.86 | Raw ScoresDC:monocyte-derived:antiCD40/VAF347: 0.19, DC:monocyte-derived:Galectin-1: 0.19, Macrophage:monocyte-derived:S._aureus: 0.18, Macrophage:Alveolar:B._anthacis_spores: 0.18, Macrophage:monocyte-derived: 0.18, DC:monocyte-derived:AEC-conditioned: 0.18, Macrophage:Alveolar: 0.18, DC:monocyte-derived:mature: 0.18, Macrophage:monocyte-derived:IFNa: 0.18, DC:monocyte-derived:CD40L: 0.18 |
| NB09_CTGAAACCAAAGCAAT-1 | Neurons:adrenal_medulla_cell_line | 0.12 | 481.85 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.27, Neuroepithelial_cell:ESC-derived: 0.26, Astrocyte:Embryonic_stem_cell-derived: 0.26, Endothelial_cells:HUVEC:VEGF: 0.25, Neurons:ES_cell-derived_neural_precursor: 0.25, Endothelial_cells:HUVEC:Serum_Amyloid_A: 0.25, Endothelial_cells:HUVEC: 0.24, Endothelial_cells:lymphatic: 0.24, Endothelial_cells:lymphatic:TNFa_48h: 0.24, Endothelial_cells:blood_vessel: 0.23 |
| NB09_GTCGTAAAGGCGTACA-1 | Macrophage:monocyte-derived:IL-4/Dex/cntrl | 0.04 | 472.58 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.24, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.23, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.23, Macrophage:monocyte-derived: 0.23, Macrophage:monocyte-derived:M-CSF: 0.23, DC:monocyte-derived:immature: 0.22, Astrocyte:Embryonic_stem_cell-derived: 0.22, Macrophage:monocyte-derived:IL-4/TGFb: 0.22, Neurons:Schwann_cell: 0.22, Macrophage:monocyte-derived:IL-4/cntrl: 0.22 |
| NB09_ACGGCCAAGCGATCCC-1 | Macrophage:monocyte-derived | 0.07 | 452.88 | Raw ScoresMacrophage:monocyte-derived: 0.22, DC:monocyte-derived: 0.22, DC:monocyte-derived:AM580: 0.22, Macrophage:monocyte-derived:IL-4/TGFb: 0.22, Macrophage:monocyte-derived:IL-4/cntrl: 0.22, DC:monocyte-derived:AEC-conditioned: 0.22, DC:monocyte-derived:immature: 0.22, Macrophage:monocyte-derived:S._aureus: 0.22, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.22, DC:monocyte-derived:anti-DC-SIGN_2h: 0.22 |
| NB09_TCAGATGAGACTACAA-1 | Neurons:adrenal_medulla_cell_line | 0.08 | 433.97 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.2, Astrocyte:Embryonic_stem_cell-derived: 0.17, Neurons:ES_cell-derived_neural_precursor: 0.16, Neuroepithelial_cell:ESC-derived: 0.16, Endothelial_cells:HUVEC:VEGF: 0.15, Macrophage:monocyte-derived:M-CSF: 0.15, iPS_cells:PDB_1lox-17Puro-10: 0.15, iPS_cells:PDB_1lox-21Puro-26: 0.15, Monocyte:MCSF: 0.15, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.15 |
| NB09_AGGGTGAGTTATCACG-1 | Macrophage:monocyte-derived | 0.10 | 394.29 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.25, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.25, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.25, DC:monocyte-derived:anti-DC-SIGN_2h: 0.24, Macrophage:monocyte-derived:IL-4/cntrl: 0.24, Macrophage:monocyte-derived:IL-4/TGFb: 0.24, Macrophage:monocyte-derived:M-CSF/IFNg: 0.23, DC:monocyte-derived: 0.23, Monocyte:leukotriene_D4: 0.23, Macrophage:monocyte-derived: 0.23 |
| NB09_GCGCAGTTCCTTAATC-1 | Neurons:adrenal_medulla_cell_line | 0.06 | 389.47 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.18, Fibroblasts:breast: 0.17, Chondrocytes:MSC-derived: 0.17, Smooth_muscle_cells:bronchial:vit_D: 0.17, Smooth_muscle_cells:bronchial: 0.17, Tissue_stem_cells:BM_MSC:TGFb3: 0.16, Osteoblasts: 0.16, Smooth_muscle_cells:vascular: 0.16, Smooth_muscle_cells:vascular:IL-17: 0.16, iPS_cells:CRL2097_foreskin: 0.16 |
| NB13_TGACGGCGTCTTGTCC-1 | Macrophage:monocyte-derived | 0.05 | 371.23 | Raw ScoresMacrophage:monocyte-derived:IFNa: 0.19, Macrophage:monocyte-derived: 0.19, Macrophage:monocyte-derived:S._aureus: 0.19, Macrophage:monocyte-derived:IL-4/cntrl: 0.19, DC:monocyte-derived:AEC-conditioned: 0.19, Macrophage:monocyte-derived:IL-4/TGFb: 0.18, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.18, Macrophage:Alveolar:B._anthacis_spores: 0.18, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.18, DC:monocyte-derived:AM580: 0.18 |
| NB09_CGATTGAGTTTGTTGG-1 | Neurons:adrenal_medulla_cell_line | 0.10 | 356.34 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.25, Neuroepithelial_cell:ESC-derived: 0.21, Astrocyte:Embryonic_stem_cell-derived: 0.2, Neurons:ES_cell-derived_neural_precursor: 0.2, iPS_cells:PDB_1lox-21Puro-26: 0.19, iPS_cells:PDB_1lox-17Puro-10: 0.19, iPS_cells:PDB_1lox-21Puro-20: 0.19, Embryonic_stem_cells: 0.19, iPS_cells:PDB_1lox-17Puro-5: 0.19, iPS_cells:PDB_2lox-22: 0.18 |
| NB09_TCACAAGAGCTAAACA-1 | Macrophage:monocyte-derived | 0.05 | 347.67 | Raw ScoresMacrophage:monocyte-derived: 0.22, Macrophage:monocyte-derived:M-CSF: 0.22, Macrophage:monocyte-derived:S._aureus: 0.22, Monocyte:MCSF: 0.22, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.22, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.22, DC:monocyte-derived: 0.22, DC:monocyte-derived:Galectin-1: 0.22, Macrophage:monocyte-derived:IFNa: 0.22, DC:monocyte-derived:immature: 0.21 |
| NB09_AAGGCAGTCTTCATGT-1 | Neurons:adrenal_medulla_cell_line | 0.08 | 340.90 | Raw ScoresAstrocyte:Embryonic_stem_cell-derived: 0.24, Neurons:adrenal_medulla_cell_line: 0.24, Neuroepithelial_cell:ESC-derived: 0.23, Neurons:ES_cell-derived_neural_precursor: 0.22, Endothelial_cells:HUVEC:VEGF: 0.21, iPS_cells:CRL2097_foreskin: 0.21, Endothelial_cells:lymphatic: 0.21, Endothelial_cells:HUVEC:Serum_Amyloid_A: 0.2, Endothelial_cells:lymphatic:TNFa_48h: 0.2, Endothelial_cells:HUVEC: 0.2 |
| NB09_TGCACCTGTAGAAGGA-1 | Macrophage:monocyte-derived | 0.02 | 323.32 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.21, Astrocyte:Embryonic_stem_cell-derived: 0.19, Neuroepithelial_cell:ESC-derived: 0.18, Macrophage:monocyte-derived: 0.17, Chondrocytes:MSC-derived: 0.17, Tissue_stem_cells:BM_MSC:BMP2: 0.17, Macrophage:monocyte-derived:S._aureus: 0.17, Tissue_stem_cells:BM_MSC:TGFb3: 0.17, Smooth_muscle_cells:vascular: 0.17, DC:monocyte-derived:mature: 0.17 |
| NB09_TAGCCGGTCACTCTTA-1 | Monocyte:CXCL4 | 0.06 | 320.68 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.2, Macrophage:monocyte-derived:M-CSF: 0.19, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.18, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.18, Macrophage:monocyte-derived: 0.18, Monocyte:CXCL4: 0.18, Monocyte:MCSF: 0.18, Macrophage:monocyte-derived:IL-4/TGFb: 0.18, Macrophage:monocyte-derived:IL-4/cntrl: 0.18, DC:monocyte-derived:anti-DC-SIGN_2h: 0.18 |
| NB09_GCAATCATCTTCAACT-1 | Neurons:adrenal_medulla_cell_line | 0.07 | 310.24 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.22, Astrocyte:Embryonic_stem_cell-derived: 0.2, Neuroepithelial_cell:ESC-derived: 0.19, Macrophage:monocyte-derived:M-CSF: 0.19, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.18, Neurons:ES_cell-derived_neural_precursor: 0.18, DC:monocyte-derived:AEC-conditioned: 0.18, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.18, DC:monocyte-derived:anti-DC-SIGN_2h: 0.18, DC:monocyte-derived: 0.17 |
| NB09_GAGGTGAGTGAAATCA-1 | Macrophage:monocyte-derived | 0.04 | 268.55 | Raw ScoresMacrophage:monocyte-derived: 0.18, Macrophage:monocyte-derived:S._aureus: 0.18, Tissue_stem_cells:BM_MSC:BMP2: 0.18, Monocyte:MCSF: 0.18, Macrophage:monocyte-derived:IFNa: 0.18, Tissue_stem_cells:BM_MSC: 0.18, Monocyte:CXCL4: 0.18, Tissue_stem_cells:BM_MSC:TGFb3: 0.18, DC:monocyte-derived:AEC-conditioned: 0.17, Tissue_stem_cells:BM_MSC:osteogenic: 0.17 |
| NB09_GAACCTAGTTGTCTTT-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.08 | 266.32 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.17, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.17, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.16, Macrophage:monocyte-derived:M-CSF: 0.16, DC:monocyte-derived:immature: 0.16, DC:monocyte-derived:Schuler_treatment: 0.16, DC:monocyte-derived: 0.16, Monocyte: 0.16, Macrophage:monocyte-derived:IL-4/cntrl: 0.16, Monocyte:S._typhimurium_flagellin: 0.16 |
| NB09_ACGGGCTGTGCGAAAC-1 | Neurons:adrenal_medulla_cell_line | 0.09 | 262.32 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.21, Astrocyte:Embryonic_stem_cell-derived: 0.18, Neuroepithelial_cell:ESC-derived: 0.18, Neurons:ES_cell-derived_neural_precursor: 0.17, iPS_cells:PDB_1lox-21Puro-26: 0.15, Embryonic_stem_cells: 0.15, iPS_cells:PDB_1lox-17Puro-10: 0.15, iPS_cells:PDB_1lox-21Puro-20: 0.15, iPS_cells:PDB_1lox-17Puro-5: 0.15, Neurons:Schwann_cell: 0.15 |
| NB09_AACTCCCAGAAGATTC-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.04 | 258.79 | Raw ScoresDC:monocyte-derived:immature: 0.15, Macrophage:monocyte-derived:M-CSF: 0.15, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.15, DC:monocyte-derived:anti-DC-SIGN_2h: 0.15, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.15, DC:monocyte-derived: 0.15, DC:monocyte-derived:Schuler_treatment: 0.15, DC:monocyte-derived:antiCD40/VAF347: 0.15, Macrophage:monocyte-derived:IL-4/TGFb: 0.15, Macrophage:monocyte-derived:IL-4/cntrl: 0.15 |
| NB09_CCAGCGAGTATCACCA-1 | Macrophage:monocyte-derived | 0.06 | 253.31 | Raw ScoresMacrophage:monocyte-derived: 0.19, Monocyte:MCSF: 0.19, Monocyte:CXCL4: 0.18, Macrophage:Alveolar: 0.18, Macrophage:monocyte-derived:S._aureus: 0.18, Macrophage:monocyte-derived:IL-4/cntrl: 0.18, Macrophage:Alveolar:B._anthacis_spores: 0.18, DC:monocyte-derived:immature: 0.18, Macrophage:monocyte-derived:IFNa: 0.18, Macrophage:monocyte-derived:IL-4/TGFb: 0.18 |
| NB09_CTTGGCTCAGCTGGCT-1 | Neurons:adrenal_medulla_cell_line | 0.02 | 246.42 | Raw ScoresAstrocyte:Embryonic_stem_cell-derived: 0.15, DC:monocyte-derived:AEC-conditioned: 0.14, Macrophage:monocyte-derived: 0.14, Macrophage:monocyte-derived:S._aureus: 0.14, Neurons:adrenal_medulla_cell_line: 0.14, DC:monocyte-derived:mature: 0.14, DC:monocyte-derived:rosiglitazone/AGN193109: 0.14, DC:monocyte-derived:AM580: 0.14, DC:monocyte-derived:immature: 0.14, Monocyte:MCSF: 0.14 |
| NB09_CTAACTTGTCACTTCC-1 | Macrophage:monocyte-derived:M-CSF | 0.08 | 242.53 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.2, DC:monocyte-derived:anti-DC-SIGN_2h: 0.2, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.19, Macrophage:monocyte-derived: 0.19, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.19, Macrophage:monocyte-derived:S._aureus: 0.19, DC:monocyte-derived:immature: 0.19, Macrophage:monocyte-derived:IFNa: 0.19, Monocyte:leukotriene_D4: 0.19, Monocyte: 0.19 |
| NB09_GATCGTAAGCTGAACG-1 | Macrophage:monocyte-derived | 0.04 | 242.07 | Raw ScoresMonocyte:MCSF: 0.18, Monocyte:CXCL4: 0.17, Macrophage:monocyte-derived: 0.17, Macrophage:monocyte-derived:S._aureus: 0.17, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.17, Macrophage:monocyte-derived:IL-4/cntrl: 0.17, Macrophage:monocyte-derived:IL-4/TGFb: 0.17, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.17, Macrophage:Alveolar: 0.17, Macrophage:monocyte-derived:IFNa: 0.16 |
| NB09_CAGAGAGCATCAGTAC-1 | Neurons:adrenal_medulla_cell_line | 0.08 | 233.23 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.2, Astrocyte:Embryonic_stem_cell-derived: 0.17, Neuroepithelial_cell:ESC-derived: 0.16, Neurons:ES_cell-derived_neural_precursor: 0.15, iPS_cells:CRL2097_foreskin: 0.14, Embryonic_stem_cells: 0.14, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.14, Macrophage:monocyte-derived:M-CSF: 0.13, iPS_cells:PDB_1lox-17Puro-5: 0.13, iPS_cells:PDB_1lox-17Puro-10: 0.13 |
| NB09_GTCTTCGTCCTCGCAT-1 | Monocyte:CXCL4 | 0.07 | 231.95 | Raw ScoresMacrophage:monocyte-derived:S._aureus: 0.18, DC:monocyte-derived:Schuler_treatment: 0.18, Macrophage:monocyte-derived:M-CSF: 0.18, Macrophage:monocyte-derived: 0.18, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.18, DC:monocyte-derived:antiCD40/VAF347: 0.18, Macrophage:Alveolar: 0.18, Macrophage:monocyte-derived:IFNa: 0.18, Macrophage:Alveolar:B._anthacis_spores: 0.18, Monocyte:CXCL4: 0.18 |
| NB09_AAGGTTCGTGCTCTTC-1 | DC:monocyte-derived:antiCD40/VAF347 | 0.08 | 224.84 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.17, Macrophage:monocyte-derived:S._aureus: 0.17, DC:monocyte-derived:Galectin-1: 0.17, DC:monocyte-derived:Schuler_treatment: 0.17, DC:monocyte-derived:antiCD40/VAF347: 0.17, DC:monocyte-derived:LPS: 0.16, DC:monocyte-derived: 0.16, Monocyte:leukotriene_D4: 0.16, Monocyte:anti-FcgRIIB: 0.16, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.16 |
| NB09_CTCGTCATCGGCGGTT-1 | Neurons:adrenal_medulla_cell_line | 0.19 | 215.55 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.27, Astrocyte:Embryonic_stem_cell-derived: 0.23, Neuroepithelial_cell:ESC-derived: 0.23, Neurons:ES_cell-derived_neural_precursor: 0.21, iPS_cells:PDB_1lox-21Puro-20: 0.19, iPS_cells:PDB_1lox-17Puro-10: 0.19, iPS_cells:PDB_1lox-21Puro-26: 0.19, iPS_cells:PDB_1lox-17Puro-5: 0.18, Embryonic_stem_cells: 0.18, iPS_cells:PDB_2lox-22: 0.18 |
| NB09_TGACTTTAGACTGGGT-1 | Neurons:adrenal_medulla_cell_line | 0.11 | 215.08 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.2, Astrocyte:Embryonic_stem_cell-derived: 0.16, Neuroepithelial_cell:ESC-derived: 0.16, Neurons:ES_cell-derived_neural_precursor: 0.15, Embryonic_stem_cells: 0.14, iPS_cells:PDB_1lox-21Puro-20: 0.13, iPS_cells:PDB_1lox-17Puro-10: 0.13, iPS_cells:PDB_1lox-17Puro-5: 0.13, iPS_cells:PDB_1lox-21Puro-26: 0.13, iPS_cells:skin_fibroblast-derived: 0.13 |
| NB09_TGAGCATGTCACACGC-1 | Neurons:adrenal_medulla_cell_line | 0.19 | 213.37 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.27, Neuroepithelial_cell:ESC-derived: 0.23, Astrocyte:Embryonic_stem_cell-derived: 0.23, Neurons:ES_cell-derived_neural_precursor: 0.22, iPS_cells:PDB_1lox-21Puro-20: 0.19, Embryonic_stem_cells: 0.19, iPS_cells:PDB_1lox-17Puro-5: 0.19, iPS_cells:PDB_1lox-17Puro-10: 0.19, iPS_cells:PDB_1lox-21Puro-26: 0.19, iPS_cells:skin_fibroblast-derived: 0.18 |
| NB14_TTTGCGCTCGAACTGT-1 | Monocyte:MCSF | 0.07 | 200.97 | Raw ScoresMacrophage:monocyte-derived:IL-4/Dex/TGFb: 0.17, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.17, Monocyte:MCSF: 0.17, Macrophage:monocyte-derived: 0.17, Macrophage:monocyte-derived:M-CSF: 0.17, Monocyte:CXCL4: 0.17, Macrophage:monocyte-derived:IL-4/TGFb: 0.17, Macrophage:monocyte-derived:IL-4/cntrl: 0.17, Macrophage:monocyte-derived:S._aureus: 0.16, Macrophage:monocyte-derived:IFNa: 0.16 |
| NB09_ACGAGCCGTTCACGGC-1 | Macrophage:monocyte-derived | 0.06 | 190.16 | Raw ScoresMacrophage:monocyte-derived: 0.2, Monocyte:MCSF: 0.2, Monocyte:CXCL4: 0.19, Macrophage:monocyte-derived:IFNa: 0.19, Macrophage:monocyte-derived:S._aureus: 0.19, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.19, Macrophage:monocyte-derived:IL-4/TGFb: 0.19, Macrophage:monocyte-derived:IL-4/cntrl: 0.19, DC:monocyte-derived:AM580: 0.19, DC:monocyte-derived:rosiglitazone: 0.19 |
| NB09_CCTTCGACATTTGCTT-1 | Neurons:adrenal_medulla_cell_line | 0.20 | 189.84 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.29, Neuroepithelial_cell:ESC-derived: 0.25, Astrocyte:Embryonic_stem_cell-derived: 0.23, Neurons:ES_cell-derived_neural_precursor: 0.22, iPS_cells:PDB_1lox-17Puro-10: 0.2, iPS_cells:PDB_1lox-21Puro-26: 0.2, Embryonic_stem_cells: 0.2, iPS_cells:PDB_1lox-21Puro-20: 0.19, iPS_cells:PDB_1lox-17Puro-5: 0.19, iPS_cells:PDB_2lox-22: 0.18 |
| NB09_CTACCCAGTGAGTATA-1 | Macrophage:monocyte-derived | 0.03 | 187.10 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.18, Monocyte:MCSF: 0.16, Macrophage:monocyte-derived: 0.16, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.16, Macrophage:monocyte-derived:M-CSF: 0.16, Neuroepithelial_cell:ESC-derived: 0.16, Monocyte:CXCL4: 0.15, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.15, Macrophage:monocyte-derived:IL-4/cntrl: 0.15, Macrophage:monocyte-derived:IL-4/TGFb: 0.15 |
| NB09_GTGCATACAGTCTTCC-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.04 | 176.19 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.17, Macrophage:monocyte-derived:M-CSF: 0.16, Astrocyte:Embryonic_stem_cell-derived: 0.16, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.16, DC:monocyte-derived:anti-DC-SIGN_2h: 0.15, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.15, Monocyte:leukotriene_D4: 0.15, Neuroepithelial_cell:ESC-derived: 0.15, DC:monocyte-derived:immature: 0.15, DC:monocyte-derived: 0.15 |
| NB09_AGCATACAGTACGCCC-1 | Neurons:adrenal_medulla_cell_line | 0.06 | 172.06 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.16, Astrocyte:Embryonic_stem_cell-derived: 0.15, Neuroepithelial_cell:ESC-derived: 0.13, Neurons:ES_cell-derived_neural_precursor: 0.13, Embryonic_stem_cells: 0.12, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.11, Smooth_muscle_cells:bronchial: 0.11, Smooth_muscle_cells:bronchial:vit_D: 0.11, DC:monocyte-derived:anti-DC-SIGN_2h: 0.11, iPS_cells:PDB_1lox-21Puro-20: 0.11 |
| NB09_CAGCTGGAGTTGAGTA-1 | Neurons:adrenal_medulla_cell_line | 0.06 | 170.80 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.15, Neuroepithelial_cell:ESC-derived: 0.13, Astrocyte:Embryonic_stem_cell-derived: 0.13, Neurons:ES_cell-derived_neural_precursor: 0.12, iPS_cells:CRL2097_foreskin: 0.12, Endothelial_cells:HUVEC:VEGF: 0.11, MSC: 0.11, Embryonic_stem_cells: 0.11, Endothelial_cells:HUVEC: 0.11, iPS_cells:skin_fibroblast: 0.11 |
| NB09_GCCTCTATCATGCATG-1 | Neurons:adrenal_medulla_cell_line | 0.10 | 169.35 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.18, Astrocyte:Embryonic_stem_cell-derived: 0.15, Neuroepithelial_cell:ESC-derived: 0.15, Neurons:ES_cell-derived_neural_precursor: 0.14, iPS_cells:PDB_1lox-17Puro-5: 0.13, iPS_cells:PDB_1lox-17Puro-10: 0.13, iPS_cells:PDB_1lox-21Puro-20: 0.13, iPS_cells:PDB_1lox-21Puro-26: 0.12, Embryonic_stem_cells: 0.12, Neurons:Schwann_cell: 0.12 |
| NB09_AGATTGCCACACCGCA-1 | Neurons:adrenal_medulla_cell_line | 0.21 | 168.21 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.31, Neuroepithelial_cell:ESC-derived: 0.27, Astrocyte:Embryonic_stem_cell-derived: 0.27, Neurons:ES_cell-derived_neural_precursor: 0.25, iPS_cells:PDB_1lox-17Puro-10: 0.23, iPS_cells:PDB_1lox-21Puro-20: 0.23, iPS_cells:PDB_1lox-17Puro-5: 0.23, iPS_cells:PDB_1lox-21Puro-26: 0.23, Embryonic_stem_cells: 0.23, iPS_cells:PDB_2lox-22: 0.22 |
| NB09_TCAGGTAAGATCACGG-1 | Tissue_stem_cells:BM_MSC:TGFb3 | 0.04 | 163.20 | Raw ScoresTissue_stem_cells:BM_MSC:TGFb3: 0.16, Tissue_stem_cells:BM_MSC:BMP2: 0.16, Smooth_muscle_cells:vascular: 0.15, DC:monocyte-derived:mature: 0.15, Fibroblasts:breast: 0.15, iPS_cells:adipose_stem_cells: 0.15, Tissue_stem_cells:BM_MSC: 0.15, DC:monocyte-derived:Schuler_treatment: 0.15, Tissue_stem_cells:BM_MSC:osteogenic: 0.15, Smooth_muscle_cells:vascular:IL-17: 0.15 |
| NB09_TGCTGCTAGTTATCGC-1 | Neurons:adrenal_medulla_cell_line | 0.13 | 156.26 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.21, Astrocyte:Embryonic_stem_cell-derived: 0.18, Neuroepithelial_cell:ESC-derived: 0.17, Neurons:ES_cell-derived_neural_precursor: 0.17, iPS_cells:PDB_1lox-21Puro-20: 0.16, iPS_cells:PDB_1lox-17Puro-5: 0.16, iPS_cells:PDB_1lox-17Puro-10: 0.15, iPS_cells:PDB_1lox-21Puro-26: 0.15, Embryonic_stem_cells: 0.15, iPS_cells:skin_fibroblast-derived: 0.14 |
| NB09_ACGCCAGTCTCTGCTG-1 | Neurons:adrenal_medulla_cell_line | 0.07 | 155.37 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.12, Astrocyte:Embryonic_stem_cell-derived: 0.1, Neuroepithelial_cell:ESC-derived: 0.1, Neurons:ES_cell-derived_neural_precursor: 0.1, Embryonic_stem_cells: 0.08, iPS_cells:PDB_1lox-17Puro-10: 0.08, Macrophage:monocyte-derived: 0.08, iPS_cells:skin_fibroblast-derived: 0.08, iPS_cells:PDB_1lox-21Puro-20: 0.08, iPS_cells:PDB_1lox-21Puro-26: 0.08 |
| NB09_GGGAGATGTGTCTGAT-1 | Monocyte:CXCL4 | 0.04 | 152.55 | Raw ScoresMacrophage:monocyte-derived:IL-4/Dex/TGFb: 0.15, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.14, Macrophage:monocyte-derived: 0.14, DC:monocyte-derived:Schuler_treatment: 0.14, Macrophage:monocyte-derived:IL-4/cntrl: 0.14, DC:monocyte-derived:AM580: 0.14, DC:monocyte-derived: 0.14, Macrophage:monocyte-derived:IL-4/TGFb: 0.14, DC:monocyte-derived:rosiglitazone: 0.14, DC:monocyte-derived:CD40L: 0.14 |
| NB09_CACAGTAGTCATCGGC-1 | Macrophage:monocyte-derived:M-CSF | 0.06 | 152.43 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.17, DC:monocyte-derived:antiCD40/VAF347: 0.17, Monocyte:S._typhimurium_flagellin: 0.17, Macrophage:monocyte-derived:M-CSF: 0.17, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.17, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.17, DC:monocyte-derived: 0.17, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.17, DC:monocyte-derived:immature: 0.16, Monocyte:anti-FcgRIIB: 0.16 |
| NB09_CTGATCCGTCCTCCAT-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.08 | 148.74 | Raw ScoresMacrophage:monocyte-derived:IL-4/Dex/TGFb: 0.19, Macrophage:monocyte-derived:M-CSF: 0.19, Macrophage:monocyte-derived:S._aureus: 0.19, DC:monocyte-derived:LPS: 0.18, Macrophage:monocyte-derived: 0.18, Macrophage:monocyte-derived:IL-4/cntrl: 0.18, Macrophage:monocyte-derived:IL-4/TGFb: 0.18, DC:monocyte-derived:Galectin-1: 0.18, Macrophage:Alveolar: 0.18, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.18 |
| NB09_AAGACCTGTTCAACCA-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 147.67 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.26, Neuroepithelial_cell:ESC-derived: 0.22, Astrocyte:Embryonic_stem_cell-derived: 0.21, Neurons:ES_cell-derived_neural_precursor: 0.21, iPS_cells:PDB_1lox-21Puro-20: 0.19, iPS_cells:PDB_1lox-21Puro-26: 0.19, iPS_cells:PDB_1lox-17Puro-10: 0.19, iPS_cells:PDB_1lox-17Puro-5: 0.19, Embryonic_stem_cells: 0.19, iPS_cells:PDB_2lox-22: 0.18 |
| NB09_GCGACCACATCCTTGC-1 | Monocyte:MCSF | 0.04 | 145.94 | Raw ScoresMacrophage:monocyte-derived: 0.16, Monocyte:MCSF: 0.16, Macrophage:monocyte-derived:S._aureus: 0.16, Macrophage:monocyte-derived:M-CSF: 0.16, Monocyte:CXCL4: 0.16, Neurons:adrenal_medulla_cell_line: 0.15, Macrophage:monocyte-derived:IL-4/cntrl: 0.15, Macrophage:monocyte-derived:IFNa: 0.15, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.15, Macrophage:monocyte-derived:IL-4/TGFb: 0.15 |
| NB09_GGAGCAAGTTGTCGCG-1 | Neurons:adrenal_medulla_cell_line | 0.18 | 145.35 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.26, Neuroepithelial_cell:ESC-derived: 0.21, Astrocyte:Embryonic_stem_cell-derived: 0.21, Neurons:ES_cell-derived_neural_precursor: 0.2, iPS_cells:PDB_1lox-17Puro-10: 0.18, iPS_cells:PDB_1lox-17Puro-5: 0.18, iPS_cells:PDB_1lox-21Puro-20: 0.17, iPS_cells:PDB_1lox-21Puro-26: 0.17, Embryonic_stem_cells: 0.17, iPS_cells:PDB_2lox-22: 0.17 |
| NB06_GGAATAAGTGACGGTA-1 | Neurons:adrenal_medulla_cell_line | 0.18 | 144.53 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.33, Neuroepithelial_cell:ESC-derived: 0.32, Neurons:ES_cell-derived_neural_precursor: 0.29, Neurons:Schwann_cell: 0.28, Embryonic_stem_cells: 0.28, iPS_cells:PDB_1lox-21Puro-20: 0.27, MSC: 0.27, iPS_cells:PDB_1lox-17Puro-10: 0.27, Tissue_stem_cells:CD326-CD56+: 0.27 |
| NB09_GGCGACTGTAAATACG-1 | Neurons:adrenal_medulla_cell_line | 0.10 | 144.32 | Raw ScoresEndothelial_cells:HUVEC:VEGF: 0.27, Endothelial_cells:lymphatic: 0.27, Endothelial_cells:HUVEC:Serum_Amyloid_A: 0.27, Endothelial_cells:lymphatic:TNFa_48h: 0.26, Endothelial_cells:HUVEC: 0.26, Endothelial_cells:blood_vessel: 0.26, Endothelial_cells:HUVEC:B._anthracis_LT: 0.25, Endothelial_cells:lymphatic:KSHV: 0.25, Endothelial_cells:HUVEC:IL-1b: 0.25, Astrocyte:Embryonic_stem_cell-derived: 0.25 |
Below shows the significant enrichments of this GEP for literature curated gene lists
This data was procured from existing single cell RNA-seq maps of neuroblastoma or related relevant data.
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by the P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
M-MDSC
These marker genes were curated for MDSC subtypes as reviewed in Veglia et. al. (PMID 33526920):
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.48e-03
Mean rank of genes in gene set: 3497.13
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CD14 | 0.0064117 | 28 | GTEx | DepMap | Descartes | 0.23 | 346.68 |
| HIF1A | 0.0052630 | 46 | GTEx | DepMap | Descartes | 1.00 | 580.88 |
| CD84 | 0.0020188 | 275 | GTEx | DepMap | Descartes | 0.07 | 17.93 |
| CD36 | 0.0019174 | 301 | GTEx | DepMap | Descartes | 0.16 | 64.74 |
| VEGFA | 0.0015821 | 403 | GTEx | DepMap | Descartes | 0.23 | 47.35 |
| TNFRSF10B | 0.0010751 | 682 | GTEx | DepMap | Descartes | 0.08 | 37.38 |
| STAT3 | 0.0008034 | 970 | GTEx | DepMap | Descartes | 0.49 | 181.26 |
| CD274 | 0.0003768 | 2093 | GTEx | DepMap | Descartes | 0.01 | 6.67 |
| IL10 | 0.0003377 | 2283 | GTEx | DepMap | Descartes | 0.00 | 9.75 |
| TGFB1 | 0.0003204 | 2369 | GTEx | DepMap | Descartes | 0.11 | 77.81 |
| ARG1 | 0.0000640 | 4564 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| NOS2 | -0.0000598 | 6589 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TNF | -0.0002312 | 9841 | GTEx | DepMap | Descartes | 0.01 | 1.28 |
| IL1B | -0.0002564 | 10187 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ARG2 | -0.0004816 | 11826 | GTEx | DepMap | Descartes | 0.05 | 23.35 |
M2 Macrophage
These genes were collated from multiple sources:
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.18e-03
Mean rank of genes in gene set: 2372.29
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CD14 | 0.0064117 | 28 | GTEx | DepMap | Descartes | 0.23 | 346.68 |
| CD163 | 0.0020899 | 259 | GTEx | DepMap | Descartes | 0.06 | 33.40 |
| VEGFA | 0.0015821 | 403 | GTEx | DepMap | Descartes | 0.23 | 47.35 |
| IL10 | 0.0003377 | 2283 | GTEx | DepMap | Descartes | 0.00 | 9.75 |
| TGFB1 | 0.0003204 | 2369 | GTEx | DepMap | Descartes | 0.11 | 77.81 |
| ARG1 | 0.0000640 | 4564 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| IL13 | -0.0000644 | 6700 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
Bridge region mesenchymal-SCP transition (Olsen)
As above but for cells in the mesenchymal transitioning to SCP region:
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.37e-03
Mean rank of genes in gene set: 367.33
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| S100A10 | 0.0059398 | 39 | GTEx | DepMap | Descartes | 0.18 | 593.98 |
| B2M | 0.0051099 | 49 | GTEx | DepMap | Descartes | 2.15 | 1602.43 |
| IFITM3 | 0.0007768 | 1014 | GTEx | DepMap | Descartes | 0.15 | 383.19 |
Below shows ranks on this GEP for literature curated gene lists for large gene sets
These include those reported as mesenchymal/adrenergic by Van Groningen et al.
High ranks indicate this gene is a driver of this GEP (note these results are not ordered).
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
VanGroningen Adrenergic Genes
Adrenergic marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 8191.66
Median rank of genes in gene set: 9776
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| HK2 | 0.0051072 | 50 | GTEx | DepMap | Descartes | 0.16 | 70.90 |
| CELF2 | 0.0035526 | 100 | GTEx | DepMap | Descartes | 0.70 | 188.22 |
| RNF144A | 0.0034976 | 101 | GTEx | DepMap | Descartes | 0.96 | 327.39 |
| MIAT | 0.0026276 | 168 | GTEx | DepMap | Descartes | 1.58 | 307.58 |
| CADM1 | 0.0024818 | 181 | GTEx | DepMap | Descartes | 3.56 | 781.33 |
| RBMS3 | 0.0023204 | 205 | GTEx | DepMap | Descartes | 4.50 | 1042.24 |
| TH | 0.0020128 | 276 | GTEx | DepMap | Descartes | 1.42 | 1389.19 |
| DAPK1 | 0.0020090 | 277 | GTEx | DepMap | Descartes | 0.54 | 188.27 |
| CXCR4 | 0.0018317 | 329 | GTEx | DepMap | Descartes | 0.15 | 167.51 |
| LYN | 0.0018138 | 331 | GTEx | DepMap | Descartes | 0.20 | 92.22 |
| NELFCD | 0.0014609 | 448 | GTEx | DepMap | Descartes | 0.42 | NA |
| MYO5A | 0.0014342 | 457 | GTEx | DepMap | Descartes | 0.48 | 76.66 |
| TMOD1 | 0.0014307 | 460 | GTEx | DepMap | Descartes | 1.00 | 616.00 |
| DPYSL3 | 0.0013355 | 512 | GTEx | DepMap | Descartes | 0.91 | 278.24 |
| DNER | 0.0012494 | 556 | GTEx | DepMap | Descartes | 0.28 | 186.35 |
| NRCAM | 0.0012120 | 580 | GTEx | DepMap | Descartes | 1.03 | 288.94 |
| PIK3R1 | 0.0011990 | 591 | GTEx | DepMap | Descartes | 0.37 | 121.10 |
| SCN3A | 0.0011677 | 617 | GTEx | DepMap | Descartes | 0.95 | 195.19 |
| UCP2 | 0.0010599 | 699 | GTEx | DepMap | Descartes | 0.09 | 112.39 |
| RIMBP2 | 0.0009828 | 773 | GTEx | DepMap | Descartes | 1.18 | 366.53 |
| ATP6V1B2 | 0.0009536 | 802 | GTEx | DepMap | Descartes | 0.30 | 100.73 |
| FAM155A | 0.0007808 | 1008 | GTEx | DepMap | Descartes | 5.27 | 1042.38 |
| IGSF3 | 0.0007237 | 1097 | GTEx | DepMap | Descartes | 0.19 | 62.91 |
| OLFM1 | 0.0006957 | 1150 | GTEx | DepMap | Descartes | 0.09 | 57.00 |
| KIDINS220 | 0.0006619 | 1212 | GTEx | DepMap | Descartes | 0.86 | 204.60 |
| TIAM1 | 0.0006491 | 1234 | GTEx | DepMap | Descartes | 0.70 | 200.19 |
| EYA1 | 0.0006273 | 1290 | GTEx | DepMap | Descartes | 0.97 | 431.13 |
| RALGDS | 0.0006197 | 1307 | GTEx | DepMap | Descartes | 0.33 | 115.78 |
| CHGB | 0.0005687 | 1427 | GTEx | DepMap | Descartes | 1.03 | 797.23 |
| GCH1 | 0.0005255 | 1535 | GTEx | DepMap | Descartes | 0.39 | 246.38 |
VanGroningen Mesenchymal Genes
Mesenchymal marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.00e-20
Mean rank of genes in gene set: 4706.94
Median rank of genes in gene set: 3929
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| APOE | 0.0162972 | 3 | GTEx | DepMap | Descartes | 2.06 | 4173.35 |
| CTSB | 0.0097785 | 8 | GTEx | DepMap | Descartes | 1.21 | 829.79 |
| RAB31 | 0.0060777 | 37 | GTEx | DepMap | Descartes | 0.36 | 219.44 |
| B2M | 0.0051099 | 49 | GTEx | DepMap | Descartes | 2.15 | 1602.43 |
| PLXDC2 | 0.0050156 | 51 | GTEx | DepMap | Descartes | 1.09 | 190.33 |
| GSN | 0.0049645 | 52 | GTEx | DepMap | Descartes | 0.33 | 112.83 |
| SGK1 | 0.0047055 | 58 | GTEx | DepMap | Descartes | 0.40 | 185.34 |
| ATP2B4 | 0.0045771 | 61 | GTEx | DepMap | Descartes | 0.99 | 214.21 |
| FNDC3B | 0.0042244 | 70 | GTEx | DepMap | Descartes | 0.85 | 216.86 |
| ROBO1 | 0.0041205 | 74 | GTEx | DepMap | Descartes | 4.85 | 1277.19 |
| ANXA2 | 0.0040949 | 75 | GTEx | DepMap | Descartes | 0.43 | 276.41 |
| NPC2 | 0.0038278 | 86 | GTEx | DepMap | Descartes | 0.18 | 321.18 |
| CBLB | 0.0036777 | 92 | GTEx | DepMap | Descartes | 0.78 | 266.54 |
| LGALS1 | 0.0034159 | 105 | GTEx | DepMap | Descartes | 0.40 | 1035.16 |
| VIM | 0.0034057 | 106 | GTEx | DepMap | Descartes | 0.66 | 521.78 |
| GRN | 0.0032178 | 111 | GTEx | DepMap | Descartes | 0.30 | 285.42 |
| PAPSS2 | 0.0031211 | 118 | GTEx | DepMap | Descartes | 0.18 | 107.17 |
| ZFP36L1 | 0.0029651 | 140 | GTEx | DepMap | Descartes | 0.36 | 274.52 |
| LPP | 0.0027199 | 159 | GTEx | DepMap | Descartes | 1.20 | 130.35 |
| COL6A2 | 0.0027115 | 162 | GTEx | DepMap | Descartes | 0.51 | 285.31 |
| IQGAP2 | 0.0026136 | 170 | GTEx | DepMap | Descartes | 0.26 | 87.90 |
| ANXA5 | 0.0025844 | 173 | GTEx | DepMap | Descartes | 0.24 | 315.05 |
| LITAF | 0.0025746 | 174 | GTEx | DepMap | Descartes | 0.30 | 263.34 |
| ANXA1 | 0.0025626 | 176 | GTEx | DepMap | Descartes | 0.24 | 273.96 |
| NRP1 | 0.0024564 | 186 | GTEx | DepMap | Descartes | 0.97 | 289.20 |
| NR3C1 | 0.0023909 | 196 | GTEx | DepMap | Descartes | 0.46 | 124.71 |
| TNS1 | 0.0022746 | 216 | GTEx | DepMap | Descartes | 0.11 | 22.16 |
| PLSCR1 | 0.0021841 | 237 | GTEx | DepMap | Descartes | 0.16 | 226.29 |
| GPR137B | 0.0021529 | 241 | GTEx | DepMap | Descartes | 0.15 | 193.27 |
| SDC2 | 0.0020560 | 267 | GTEx | DepMap | Descartes | 0.14 | 63.59 |
Descartes adrenocortical markers
Top 50 marker genes of adrenocortical cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/adrenocortical/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 5.56e-01
Mean rank of genes in gene set: 6359.42
Median rank of genes in gene set: 6579.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| APOC1 | 0.0140010 | 4 | GTEx | DepMap | Descartes | 1.35 | 4965.71 |
| PAPSS2 | 0.0031211 | 118 | GTEx | DepMap | Descartes | 0.18 | 107.17 |
| SH3BP5 | 0.0017280 | 356 | GTEx | DepMap | Descartes | 0.19 | 109.91 |
| DNER | 0.0012494 | 556 | GTEx | DepMap | Descartes | 0.28 | 186.35 |
| NPC1 | 0.0009179 | 844 | GTEx | DepMap | Descartes | 0.24 | 94.68 |
| FDX1 | 0.0008848 | 878 | GTEx | DepMap | Descartes | 0.06 | 42.37 |
| SH3PXD2B | 0.0005938 | 1373 | GTEx | DepMap | Descartes | 0.19 | 43.26 |
| CLU | 0.0003594 | 2170 | GTEx | DepMap | Descartes | 0.38 | 220.10 |
| SCARB1 | 0.0002945 | 2488 | GTEx | DepMap | Descartes | 0.09 | 30.38 |
| PEG3 | 0.0002790 | 2584 | GTEx | DepMap | Descartes | 0.26 | NA |
| IGF1R | 0.0002134 | 3034 | GTEx | DepMap | Descartes | 0.77 | 102.70 |
| ERN1 | 0.0002111 | 3047 | GTEx | DepMap | Descartes | 0.06 | 12.91 |
| JAKMIP2 | 0.0001999 | 3141 | GTEx | DepMap | Descartes | 0.40 | 85.78 |
| SCAP | 0.0001326 | 3770 | GTEx | DepMap | Descartes | 0.14 | 65.95 |
| DHCR24 | 0.0000606 | 4616 | GTEx | DepMap | Descartes | 0.08 | 29.01 |
| INHA | 0.0000392 | 4911 | GTEx | DepMap | Descartes | 0.00 | 0.79 |
| GRAMD1B | -0.0000193 | 5810 | GTEx | DepMap | Descartes | 0.07 | 12.07 |
| FREM2 | -0.0000422 | 6234 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| BAIAP2L1 | -0.0000763 | 6925 | GTEx | DepMap | Descartes | 0.02 | 4.60 |
| STAR | -0.0000985 | 7374 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| POR | -0.0001309 | 8070 | GTEx | DepMap | Descartes | 0.06 | 37.80 |
| DHCR7 | -0.0001575 | 8627 | GTEx | DepMap | Descartes | 0.02 | 4.09 |
| GSTA4 | -0.0001760 | 8950 | GTEx | DepMap | Descartes | 0.14 | 145.61 |
| FDXR | -0.0001857 | 9114 | GTEx | DepMap | Descartes | 0.01 | 5.78 |
| FRMD5 | -0.0002434 | 10018 | GTEx | DepMap | Descartes | 0.75 | 263.22 |
| TM7SF2 | -0.0002480 | 10073 | GTEx | DepMap | Descartes | 0.06 | 48.21 |
| HMGCS1 | -0.0002661 | 10303 | GTEx | DepMap | Descartes | 0.09 | 26.83 |
| SLC1A2 | -0.0002752 | 10409 | GTEx | DepMap | Descartes | 0.15 | 20.91 |
| LDLR | -0.0003299 | 10966 | GTEx | DepMap | Descartes | 0.05 | 11.50 |
| HMGCR | -0.0003369 | 11020 | GTEx | DepMap | Descartes | 0.09 | 34.44 |
Descartes chromaffin markers
Top 50 marker genes of chromaffin cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/chromaffin/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.97e-01
Mean rank of genes in gene set: 7840.46
Median rank of genes in gene set: 10344
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ANKFN1 | 0.0043625 | 65 | GTEx | DepMap | Descartes | 1.30 | 618.93 |
| PLXNA4 | 0.0036461 | 95 | GTEx | DepMap | Descartes | 0.90 | 132.86 |
| SLC44A5 | 0.0024493 | 188 | GTEx | DepMap | Descartes | 1.02 | 495.14 |
| NTRK1 | 0.0023553 | 201 | GTEx | DepMap | Descartes | 0.45 | 266.60 |
| CNKSR2 | 0.0015397 | 415 | GTEx | DepMap | Descartes | 0.78 | 187.06 |
| TMEFF2 | 0.0015149 | 426 | GTEx | DepMap | Descartes | 0.41 | 279.67 |
| EYA1 | 0.0006273 | 1290 | GTEx | DepMap | Descartes | 0.97 | 431.13 |
| FAT3 | 0.0005279 | 1531 | GTEx | DepMap | Descartes | 0.35 | 30.17 |
| GAP43 | 0.0004713 | 1716 | GTEx | DepMap | Descartes | 0.76 | 712.89 |
| PTCHD1 | 0.0004257 | 1883 | GTEx | DepMap | Descartes | 0.23 | 33.94 |
| TMEM132C | 0.0004237 | 1894 | GTEx | DepMap | Descartes | 2.20 | 838.87 |
| CNTFR | 0.0003689 | 2120 | GTEx | DepMap | Descartes | 0.20 | 178.49 |
| REEP1 | 0.0002260 | 2924 | GTEx | DepMap | Descartes | 0.26 | 120.57 |
| RPH3A | 0.0000456 | 4820 | GTEx | DepMap | Descartes | 0.02 | 8.42 |
| IL7 | -0.0000818 | 7047 | GTEx | DepMap | Descartes | 0.99 | 1027.65 |
| RGMB | -0.0000881 | 7169 | GTEx | DepMap | Descartes | 0.25 | 94.03 |
| GREM1 | -0.0000927 | 7248 | GTEx | DepMap | Descartes | 0.01 | 2.20 |
| EYA4 | -0.0001126 | 7700 | GTEx | DepMap | Descartes | 0.73 | 268.72 |
| CCND1 | -0.0001303 | 8055 | GTEx | DepMap | Descartes | 1.25 | 516.27 |
| MAB21L1 | -0.0002533 | 10143 | GTEx | DepMap | Descartes | 0.11 | 56.13 |
| BASP1 | -0.0002693 | 10344 | GTEx | DepMap | Descartes | 0.52 | 486.77 |
| RBFOX1 | -0.0002883 | 10544 | GTEx | DepMap | Descartes | 3.49 | 1199.43 |
| KCNB2 | -0.0003183 | 10860 | GTEx | DepMap | Descartes | 0.66 | 314.64 |
| SLC6A2 | -0.0003806 | 11372 | GTEx | DepMap | Descartes | 0.13 | 65.49 |
| SYNPO2 | -0.0005391 | 12007 | GTEx | DepMap | Descartes | 0.25 | 24.58 |
| MAB21L2 | -0.0007266 | 12315 | GTEx | DepMap | Descartes | 0.02 | 10.27 |
| RYR2 | -0.0007920 | 12363 | GTEx | DepMap | Descartes | 2.13 | 212.40 |
| EPHA6 | -0.0008785 | 12411 | GTEx | DepMap | Descartes | 0.23 | 96.58 |
| ELAVL2 | -0.0009034 | 12422 | GTEx | DepMap | Descartes | 0.59 | 230.68 |
| TUBB2A | -0.0009071 | 12424 | GTEx | DepMap | Descartes | 0.30 | 245.66 |
Descartes Vascular_endothelial markers
Top 50 marker genes of Vascular_endothelial cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/vascular_endothelial/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 8434.92
Median rank of genes in gene set: 9006.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CDH13 | 0.0029511 | 142 | GTEx | DepMap | Descartes | 0.64 | 166.78 |
| EHD3 | 0.0005995 | 1358 | GTEx | DepMap | Descartes | 0.05 | 21.80 |
| CYP26B1 | 0.0003169 | 2386 | GTEx | DepMap | Descartes | 0.03 | 4.63 |
| CRHBP | 0.0001230 | 3892 | GTEx | DepMap | Descartes | 0.00 | 2.37 |
| RASIP1 | 0.0001022 | 4114 | GTEx | DepMap | Descartes | 0.03 | 25.19 |
| KANK3 | 0.0000954 | 4198 | GTEx | DepMap | Descartes | 0.02 | 10.73 |
| HYAL2 | 0.0000381 | 4930 | GTEx | DepMap | Descartes | 0.04 | 14.01 |
| CALCRL | 0.0000339 | 4996 | GTEx | DepMap | Descartes | 0.14 | 20.62 |
| PLVAP | 0.0000002 | 5499 | GTEx | DepMap | Descartes | 0.05 | 35.85 |
| TMEM88 | -0.0000566 | 6537 | GTEx | DepMap | Descartes | 0.06 | 116.71 |
| TIE1 | -0.0000602 | 6603 | GTEx | DepMap | Descartes | 0.02 | 5.17 |
| MMRN2 | -0.0000952 | 7294 | GTEx | DepMap | Descartes | 0.01 | 1.79 |
| GALNT15 | -0.0000992 | 7389 | GTEx | DepMap | Descartes | 0.01 | NA |
| NPR1 | -0.0001324 | 8101 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| RAMP2 | -0.0001336 | 8133 | GTEx | DepMap | Descartes | 0.06 | 96.36 |
| CDH5 | -0.0001412 | 8284 | GTEx | DepMap | Descartes | 0.03 | 4.93 |
| F8 | -0.0001451 | 8373 | GTEx | DepMap | Descartes | 0.00 | 0.12 |
| CLDN5 | -0.0001473 | 8428 | GTEx | DepMap | Descartes | 0.03 | 6.49 |
| SLCO2A1 | -0.0001726 | 8901 | GTEx | DepMap | Descartes | 0.02 | 2.20 |
| IRX3 | -0.0001857 | 9112 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| NR5A2 | -0.0001948 | 9268 | GTEx | DepMap | Descartes | 0.06 | 15.93 |
| CEACAM1 | -0.0002409 | 9980 | GTEx | DepMap | Descartes | 0.02 | 17.49 |
| FLT4 | -0.0002474 | 10055 | GTEx | DepMap | Descartes | 0.01 | 2.81 |
| TEK | -0.0002529 | 10139 | GTEx | DepMap | Descartes | 0.03 | 5.46 |
| PODXL | -0.0002961 | 10642 | GTEx | DepMap | Descartes | 0.08 | 20.40 |
| MYRIP | -0.0003110 | 10784 | GTEx | DepMap | Descartes | 0.08 | 22.05 |
| SHE | -0.0003171 | 10839 | GTEx | DepMap | Descartes | 0.02 | 5.02 |
| SHANK3 | -0.0003788 | 11356 | GTEx | DepMap | Descartes | 0.06 | 8.87 |
| ROBO4 | -0.0003802 | 11369 | GTEx | DepMap | Descartes | 0.01 | 4.66 |
| KDR | -0.0003848 | 11398 | GTEx | DepMap | Descartes | 0.01 | 0.34 |
Descartes stromal markers
Top 50 marker genes of stromal cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/stromal/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 6.91e-01
Mean rank of genes in gene set: 6546.75
Median rank of genes in gene set: 7082.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| PRICKLE1 | 0.0019651 | 292 | GTEx | DepMap | Descartes | 0.67 | 240.72 |
| IGFBP3 | 0.0007493 | 1058 | GTEx | DepMap | Descartes | 0.20 | 51.13 |
| COL6A3 | 0.0007060 | 1121 | GTEx | DepMap | Descartes | 0.23 | 41.50 |
| DKK2 | 0.0005622 | 1444 | GTEx | DepMap | Descartes | 0.04 | 22.77 |
| EDNRA | 0.0004672 | 1728 | GTEx | DepMap | Descartes | 0.08 | 29.48 |
| ADAMTS2 | 0.0004070 | 1965 | GTEx | DepMap | Descartes | 0.16 | 36.27 |
| COL3A1 | 0.0004028 | 1978 | GTEx | DepMap | Descartes | 0.55 | 197.24 |
| ELN | 0.0003535 | 2200 | GTEx | DepMap | Descartes | 0.15 | 60.10 |
| ABCC9 | 0.0003452 | 2237 | GTEx | DepMap | Descartes | 0.09 | 14.36 |
| CCDC80 | 0.0003422 | 2248 | GTEx | DepMap | Descartes | 0.07 | 11.78 |
| COL1A2 | 0.0002710 | 2634 | GTEx | DepMap | Descartes | 0.84 | 243.19 |
| PRRX1 | 0.0002639 | 2678 | GTEx | DepMap | Descartes | 0.05 | 16.38 |
| PAMR1 | 0.0002555 | 2719 | GTEx | DepMap | Descartes | 0.07 | 41.58 |
| POSTN | 0.0002400 | 2828 | GTEx | DepMap | Descartes | 0.16 | 74.33 |
| C7 | 0.0001315 | 3785 | GTEx | DepMap | Descartes | 0.03 | 5.53 |
| DCN | 0.0001243 | 3876 | GTEx | DepMap | Descartes | 0.05 | 15.63 |
| ITGA11 | 0.0000368 | 4948 | GTEx | DepMap | Descartes | 0.04 | 6.74 |
| CD248 | 0.0000345 | 4989 | GTEx | DepMap | Descartes | 0.02 | 13.10 |
| LAMC3 | 0.0000226 | 5160 | GTEx | DepMap | Descartes | 0.00 | 0.84 |
| COL27A1 | 0.0000059 | 5415 | GTEx | DepMap | Descartes | 0.05 | 15.06 |
| LOX | -0.0000549 | 6501 | GTEx | DepMap | Descartes | 0.02 | 6.23 |
| SCARA5 | -0.0000803 | 7013 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| COL1A1 | -0.0000871 | 7152 | GTEx | DepMap | Descartes | 0.65 | 163.90 |
| GLI2 | -0.0001013 | 7425 | GTEx | DepMap | Descartes | 0.03 | 8.51 |
| SFRP2 | -0.0001057 | 7523 | GTEx | DepMap | Descartes | 0.04 | 52.67 |
| RSPO3 | -0.0001116 | 7670 | GTEx | DepMap | Descartes | 0.00 | NA |
| ADAMTSL3 | -0.0001165 | 7779 | GTEx | DepMap | Descartes | 0.02 | 3.09 |
| ISLR | -0.0001483 | 8446 | GTEx | DepMap | Descartes | 0.01 | 1.44 |
| ACTA2 | -0.0001799 | 9010 | GTEx | DepMap | Descartes | 0.07 | 57.83 |
| OGN | -0.0001855 | 9101 | GTEx | DepMap | Descartes | 0.01 | 5.31 |
Descartes sympathoblasts markers
Top 50 marker genes of sympathoblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/sympathoblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 4.49e-01
Mean rank of genes in gene set: 6200.13
Median rank of genes in gene set: 5232.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ROBO1 | 0.0041205 | 74 | GTEx | DepMap | Descartes | 4.85 | 1277.19 |
| MGAT4C | 0.0033319 | 108 | GTEx | DepMap | Descartes | 0.93 | 77.31 |
| GRID2 | 0.0027552 | 158 | GTEx | DepMap | Descartes | 1.00 | 355.15 |
| NTNG1 | 0.0012849 | 539 | GTEx | DepMap | Descartes | 0.74 | 311.70 |
| UNC80 | 0.0012748 | 542 | GTEx | DepMap | Descartes | 0.98 | 120.38 |
| PCSK2 | 0.0010565 | 701 | GTEx | DepMap | Descartes | 0.33 | 147.51 |
| ST18 | 0.0009347 | 822 | GTEx | DepMap | Descartes | 0.14 | 46.53 |
| TENM1 | 0.0009142 | 849 | GTEx | DepMap | Descartes | 0.54 | NA |
| DGKK | 0.0009090 | 853 | GTEx | DepMap | Descartes | 0.04 | 13.90 |
| SLC18A1 | 0.0008917 | 872 | GTEx | DepMap | Descartes | 0.23 | 157.78 |
| FAM155A | 0.0007808 | 1008 | GTEx | DepMap | Descartes | 5.27 | 1042.38 |
| TIAM1 | 0.0006491 | 1234 | GTEx | DepMap | Descartes | 0.70 | 200.19 |
| LAMA3 | 0.0006266 | 1292 | GTEx | DepMap | Descartes | 0.12 | 19.15 |
| SLC24A2 | 0.0005783 | 1405 | GTEx | DepMap | Descartes | 0.09 | 15.55 |
| CHGB | 0.0005687 | 1427 | GTEx | DepMap | Descartes | 1.03 | 797.23 |
| GCH1 | 0.0005255 | 1535 | GTEx | DepMap | Descartes | 0.39 | 246.38 |
| AGBL4 | 0.0004242 | 1890 | GTEx | DepMap | Descartes | 1.48 | 589.76 |
| CNTN3 | 0.0002135 | 3030 | GTEx | DepMap | Descartes | 0.07 | 26.75 |
| HTATSF1 | 0.0000275 | 5100 | GTEx | DepMap | Descartes | 0.09 | 58.51 |
| SPOCK3 | 0.0000086 | 5365 | GTEx | DepMap | Descartes | 0.22 | 148.14 |
| TBX20 | -0.0000203 | 5827 | GTEx | DepMap | Descartes | 0.12 | 120.73 |
| PENK | -0.0001229 | 7903 | GTEx | DepMap | Descartes | 0.00 | 3.33 |
| SORCS3 | -0.0002274 | 9787 | GTEx | DepMap | Descartes | 0.01 | 1.74 |
| C1QL1 | -0.0003618 | 11223 | GTEx | DepMap | Descartes | 0.01 | 5.41 |
| KCTD16 | -0.0004691 | 11788 | GTEx | DepMap | Descartes | 0.69 | 72.03 |
| GRM7 | -0.0004949 | 11866 | GTEx | DepMap | Descartes | 0.07 | 26.87 |
| PACRG | -0.0005165 | 11934 | GTEx | DepMap | Descartes | 0.20 | 219.54 |
| ARC | -0.0005395 | 12008 | GTEx | DepMap | Descartes | 0.02 | 8.97 |
| SLC35F3 | -0.0006673 | 12244 | GTEx | DepMap | Descartes | 0.06 | 19.10 |
| CHGA | -0.0007719 | 12350 | GTEx | DepMap | Descartes | 0.56 | 280.95 |
Descartes erythroblasts markers
Top 50 marker genes of erythroblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/erythroblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 5.03e-02
Mean rank of genes in gene set: 5171.38
Median rank of genes in gene set: 4045
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| BLVRB | 0.0030002 | 136 | GTEx | DepMap | Descartes | 0.10 | 146.71 |
| SPECC1 | 0.0011398 | 638 | GTEx | DepMap | Descartes | 0.07 | 16.43 |
| DENND4A | 0.0008069 | 965 | GTEx | DepMap | Descartes | 0.64 | 123.34 |
| SNCA | 0.0005707 | 1423 | GTEx | DepMap | Descartes | 0.14 | 72.06 |
| GYPC | 0.0005409 | 1504 | GTEx | DepMap | Descartes | 0.01 | 7.52 |
| MICAL2 | 0.0004927 | 1642 | GTEx | DepMap | Descartes | 0.08 | 28.75 |
| EPB41 | 0.0004205 | 1909 | GTEx | DepMap | Descartes | 0.44 | 130.68 |
| SLC25A21 | 0.0002772 | 2594 | GTEx | DepMap | Descartes | 0.02 | 17.37 |
| SLC25A37 | 0.0002755 | 2605 | GTEx | DepMap | Descartes | 0.10 | 49.44 |
| TRAK2 | 0.0002143 | 3021 | GTEx | DepMap | Descartes | 0.10 | 26.46 |
| XPO7 | 0.0002138 | 3029 | GTEx | DepMap | Descartes | 0.20 | 73.17 |
| SPTB | 0.0001814 | 3303 | GTEx | DepMap | Descartes | 0.08 | 11.28 |
| MARCH3 | 0.0001621 | 3480 | GTEx | DepMap | Descartes | 0.16 | NA |
| CAT | 0.0001119 | 4003 | GTEx | DepMap | Descartes | 0.05 | 39.81 |
| CPOX | 0.0001081 | 4045 | GTEx | DepMap | Descartes | 0.03 | 24.93 |
| ABCB10 | 0.0000388 | 4918 | GTEx | DepMap | Descartes | 0.09 | 41.83 |
| RGS6 | 0.0000081 | 5373 | GTEx | DepMap | Descartes | 0.01 | 2.74 |
| TMCC2 | 0.0000039 | 5444 | GTEx | DepMap | Descartes | 0.04 | 14.32 |
| SELENBP1 | -0.0000292 | 5982 | GTEx | DepMap | Descartes | 0.00 | 4.14 |
| ANK1 | -0.0000428 | 6247 | GTEx | DepMap | Descartes | 0.16 | 34.44 |
| RAPGEF2 | -0.0000607 | 6613 | GTEx | DepMap | Descartes | 0.40 | 72.44 |
| ALAS2 | -0.0000995 | 7395 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SLC4A1 | -0.0001276 | 8010 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| GCLC | -0.0001475 | 8431 | GTEx | DepMap | Descartes | 0.08 | 50.77 |
| FECH | -0.0002914 | 10587 | GTEx | DepMap | Descartes | 0.03 | 5.43 |
| TSPAN5 | -0.0002924 | 10601 | GTEx | DepMap | Descartes | 0.46 | 193.80 |
| RHD | -0.0003642 | 11245 | GTEx | DepMap | Descartes | 0.01 | 2.31 |
| TFR2 | -0.0007900 | 12359 | GTEx | DepMap | Descartes | 0.09 | 34.49 |
| SOX6 | -0.0010422 | 12468 | GTEx | DepMap | Descartes | 0.34 | 49.81 |
| NA | NA | NA | NA | NA | NA | NA | NA |
Descartes myeloid markers
Top 50 marker genes of myeloid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/myeloid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 7.00e-15
Mean rank of genes in gene set: 1758.76
Median rank of genes in gene set: 344
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| SPP1 | 0.0298595 | 1 | GTEx | DepMap | Descartes | 11.14 | 16904.96 |
| CTSD | 0.0133510 | 5 | GTEx | DepMap | Descartes | 1.66 | 2008.08 |
| CTSB | 0.0097785 | 8 | GTEx | DepMap | Descartes | 1.21 | 829.79 |
| CD14 | 0.0064117 | 28 | GTEx | DepMap | Descartes | 0.23 | 346.68 |
| TGFBI | 0.0061742 | 32 | GTEx | DepMap | Descartes | 0.32 | 164.77 |
| ABCA1 | 0.0047091 | 57 | GTEx | DepMap | Descartes | 0.46 | 98.15 |
| LGMN | 0.0039473 | 80 | GTEx | DepMap | Descartes | 0.42 | 475.11 |
| HRH1 | 0.0036414 | 96 | GTEx | DepMap | Descartes | 0.25 | 126.32 |
| FMN1 | 0.0033496 | 107 | GTEx | DepMap | Descartes | 2.15 | 313.62 |
| SFMBT2 | 0.0031337 | 115 | GTEx | DepMap | Descartes | 0.69 | 158.28 |
| MSR1 | 0.0030759 | 124 | GTEx | DepMap | Descartes | 0.22 | 163.99 |
| SLC1A3 | 0.0026384 | 167 | GTEx | DepMap | Descartes | 0.15 | 91.50 |
| ATP8B4 | 0.0024488 | 189 | GTEx | DepMap | Descartes | 0.10 | 41.00 |
| MERTK | 0.0022964 | 210 | GTEx | DepMap | Descartes | 0.13 | 73.34 |
| MS4A4A | 0.0022004 | 234 | GTEx | DepMap | Descartes | 0.04 | 55.21 |
| CD163 | 0.0020899 | 259 | GTEx | DepMap | Descartes | 0.06 | 33.40 |
| CTSS | 0.0019710 | 291 | GTEx | DepMap | Descartes | 0.11 | 57.30 |
| CST3 | 0.0018489 | 321 | GTEx | DepMap | Descartes | 0.31 | 192.93 |
| RBPJ | 0.0018444 | 325 | GTEx | DepMap | Descartes | 0.68 | 309.73 |
| CSF1R | 0.0017096 | 363 | GTEx | DepMap | Descartes | 0.08 | 42.15 |
| PTPRE | 0.0013545 | 504 | GTEx | DepMap | Descartes | 0.23 | 76.40 |
| CD74 | 0.0012356 | 562 | GTEx | DepMap | Descartes | 0.48 | 320.12 |
| SLCO2B1 | 0.0011583 | 622 | GTEx | DepMap | Descartes | 0.13 | 55.78 |
| FGL2 | 0.0010673 | 689 | GTEx | DepMap | Descartes | 0.03 | 12.90 |
| FGD2 | 0.0010120 | 741 | GTEx | DepMap | Descartes | 0.07 | 23.82 |
| ADAP2 | 0.0008760 | 890 | GTEx | DepMap | Descartes | 0.16 | 111.47 |
| CPVL | 0.0008467 | 928 | GTEx | DepMap | Descartes | 0.09 | 156.20 |
| CTSC | 0.0007160 | 1109 | GTEx | DepMap | Descartes | 0.10 | 33.82 |
| CYBB | 0.0007067 | 1119 | GTEx | DepMap | Descartes | 0.07 | 25.85 |
| ITPR2 | 0.0005740 | 1413 | GTEx | DepMap | Descartes | 0.29 | 49.44 |
Descartes Schwann markers
Top 50 marker genes of Schwann cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/schwann/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 7.78e-02
Mean rank of genes in gene set: 5500.82
Median rank of genes in gene set: 5393.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TRPM3 | 0.0070133 | 25 | GTEx | DepMap | Descartes | 0.95 | 166.63 |
| VIM | 0.0034057 | 106 | GTEx | DepMap | Descartes | 0.66 | 521.78 |
| LRRTM4 | 0.0024769 | 182 | GTEx | DepMap | Descartes | 2.39 | 1322.71 |
| GAS7 | 0.0020198 | 274 | GTEx | DepMap | Descartes | 0.22 | 65.49 |
| NRXN3 | 0.0016294 | 388 | GTEx | DepMap | Descartes | 0.49 | 117.97 |
| SCN7A | 0.0015865 | 401 | GTEx | DepMap | Descartes | 0.34 | 91.44 |
| FIGN | 0.0015837 | 402 | GTEx | DepMap | Descartes | 0.40 | 87.74 |
| PPP2R2B | 0.0014896 | 438 | GTEx | DepMap | Descartes | 0.88 | 157.74 |
| STARD13 | 0.0013958 | 480 | GTEx | DepMap | Descartes | 0.23 | 91.87 |
| PMP22 | 0.0012151 | 576 | GTEx | DepMap | Descartes | 0.50 | 560.84 |
| DST | 0.0010478 | 710 | GTEx | DepMap | Descartes | 2.05 | 179.21 |
| COL18A1 | 0.0010007 | 754 | GTEx | DepMap | Descartes | 0.43 | 99.95 |
| VCAN | 0.0007997 | 978 | GTEx | DepMap | Descartes | 0.29 | 54.23 |
| SFRP1 | 0.0006363 | 1267 | GTEx | DepMap | Descartes | 0.15 | 84.96 |
| NRXN1 | 0.0005434 | 1489 | GTEx | DepMap | Descartes | 1.50 | 321.37 |
| GRIK3 | 0.0005109 | 1580 | GTEx | DepMap | Descartes | 0.05 | 8.97 |
| KCTD12 | 0.0003502 | 2210 | GTEx | DepMap | Descartes | 0.05 | 14.46 |
| PTN | 0.0002330 | 2875 | GTEx | DepMap | Descartes | 0.19 | 199.04 |
| PTPRZ1 | 0.0002191 | 2992 | GTEx | DepMap | Descartes | 0.00 | 2.73 |
| SLC35F1 | 0.0001793 | 3320 | GTEx | DepMap | Descartes | 0.04 | 16.99 |
| LAMC1 | 0.0000648 | 4552 | GTEx | DepMap | Descartes | 0.16 | 40.14 |
| OLFML2A | 0.0000115 | 5309 | GTEx | DepMap | Descartes | 0.01 | 1.01 |
| MDGA2 | 0.0000023 | 5478 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| LAMA4 | -0.0000103 | 5667 | GTEx | DepMap | Descartes | 0.08 | 18.39 |
| XKR4 | -0.0000187 | 5794 | GTEx | DepMap | Descartes | 0.18 | 14.92 |
| MPZ | -0.0000244 | 5896 | GTEx | DepMap | Descartes | 0.00 | 0.48 |
| IL1RAPL2 | -0.0000277 | 5953 | GTEx | DepMap | Descartes | 0.04 | 21.39 |
| HMGA2 | -0.0000791 | 6980 | GTEx | DepMap | Descartes | 0.01 | 3.27 |
| PLP1 | -0.0001275 | 8007 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ERBB3 | -0.0001355 | 8171 | GTEx | DepMap | Descartes | 0.01 | 4.29 |
Descartes Megakaryocytes markers
Top 50 marker genes of Megakaryocytes cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/megakaryocytes/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 6.57e-06
Mean rank of genes in gene set: 3925.76
Median rank of genes in gene set: 2369
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| SLC2A3 | 0.0092391 | 11 | GTEx | DepMap | Descartes | 0.64 | 323.63 |
| GSN | 0.0049645 | 52 | GTEx | DepMap | Descartes | 0.33 | 112.83 |
| CD9 | 0.0028476 | 148 | GTEx | DepMap | Descartes | 0.28 | 364.10 |
| LIMS1 | 0.0026864 | 164 | GTEx | DepMap | Descartes | 0.44 | 215.66 |
| PLEK | 0.0021910 | 236 | GTEx | DepMap | Descartes | 0.07 | 54.40 |
| CD84 | 0.0020188 | 275 | GTEx | DepMap | Descartes | 0.07 | 17.93 |
| PDE3A | 0.0018899 | 309 | GTEx | DepMap | Descartes | 1.09 | 277.90 |
| FERMT3 | 0.0017909 | 341 | GTEx | DepMap | Descartes | 0.05 | 51.74 |
| ACTB | 0.0017595 | 348 | GTEx | DepMap | Descartes | 3.15 | 2544.66 |
| UBASH3B | 0.0017183 | 359 | GTEx | DepMap | Descartes | 0.15 | 43.48 |
| FLNA | 0.0010414 | 717 | GTEx | DepMap | Descartes | 0.32 | 70.17 |
| MYH9 | 0.0009450 | 812 | GTEx | DepMap | Descartes | 0.31 | 62.60 |
| TUBB1 | 0.0009260 | 833 | GTEx | DepMap | Descartes | 0.01 | 5.68 |
| RAP1B | 0.0007863 | 1004 | GTEx | DepMap | Descartes | 0.25 | 35.93 |
| ZYX | 0.0006346 | 1273 | GTEx | DepMap | Descartes | 0.10 | 111.64 |
| TPM4 | 0.0006122 | 1323 | GTEx | DepMap | Descartes | 0.21 | 93.94 |
| ACTN1 | 0.0005739 | 1415 | GTEx | DepMap | Descartes | 0.33 | 136.97 |
| SPN | 0.0005662 | 1433 | GTEx | DepMap | Descartes | 0.02 | 3.77 |
| TMSB4X | 0.0005611 | 1447 | GTEx | DepMap | Descartes | 3.49 | 2955.18 |
| FLI1 | 0.0005206 | 1552 | GTEx | DepMap | Descartes | 0.16 | 52.34 |
| MCTP1 | 0.0004019 | 1986 | GTEx | DepMap | Descartes | 0.39 | 86.15 |
| INPP4B | 0.0003804 | 2070 | GTEx | DepMap | Descartes | 0.59 | 130.07 |
| TGFB1 | 0.0003204 | 2369 | GTEx | DepMap | Descartes | 0.11 | 77.81 |
| TLN1 | 0.0003138 | 2402 | GTEx | DepMap | Descartes | 0.09 | 16.03 |
| P2RX1 | 0.0002655 | 2667 | GTEx | DepMap | Descartes | 0.01 | 9.14 |
| STON2 | 0.0002538 | 2733 | GTEx | DepMap | Descartes | 0.07 | 36.92 |
| STOM | 0.0002379 | 2839 | GTEx | DepMap | Descartes | 0.08 | 47.84 |
| PSTPIP2 | 0.0001492 | 3599 | GTEx | DepMap | Descartes | 0.02 | 15.18 |
| THBS1 | 0.0001466 | 3631 | GTEx | DepMap | Descartes | 0.19 | 63.22 |
| ARHGAP6 | 0.0001258 | 3850 | GTEx | DepMap | Descartes | 0.06 | 17.31 |
Descartes Lyphoid markers
Top 50 marker genes of Lyphoid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/lymphoid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.17e-05
Mean rank of genes in gene set: 4187.36
Median rank of genes in gene set: 1728.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| B2M | 0.0051099 | 49 | GTEx | DepMap | Descartes | 2.15 | 1602.43 |
| ITPKB | 0.0035924 | 98 | GTEx | DepMap | Descartes | 0.16 | 66.72 |
| CELF2 | 0.0035526 | 100 | GTEx | DepMap | Descartes | 0.70 | 188.22 |
| MSN | 0.0022112 | 232 | GTEx | DepMap | Descartes | 0.16 | 74.05 |
| ARID5B | 0.0021522 | 242 | GTEx | DepMap | Descartes | 0.30 | 83.20 |
| LCP1 | 0.0018934 | 306 | GTEx | DepMap | Descartes | 0.10 | 61.02 |
| CD44 | 0.0017946 | 338 | GTEx | DepMap | Descartes | 0.71 | 274.79 |
| PTPRC | 0.0017466 | 350 | GTEx | DepMap | Descartes | 0.19 | 99.90 |
| STK39 | 0.0017006 | 366 | GTEx | DepMap | Descartes | 0.89 | 519.27 |
| PLEKHA2 | 0.0015445 | 413 | GTEx | DepMap | Descartes | 0.14 | 58.64 |
| ARHGDIB | 0.0011735 | 615 | GTEx | DepMap | Descartes | 0.05 | 67.39 |
| RAP1GAP2 | 0.0008721 | 898 | GTEx | DepMap | Descartes | 1.09 | 287.30 |
| PRKCH | 0.0008604 | 912 | GTEx | DepMap | Descartes | 0.20 | 97.75 |
| GNG2 | 0.0008202 | 949 | GTEx | DepMap | Descartes | 0.50 | 265.83 |
| FOXP1 | 0.0007875 | 1001 | GTEx | DepMap | Descartes | 1.45 | 288.35 |
| SP100 | 0.0007125 | 1114 | GTEx | DepMap | Descartes | 0.12 | 46.33 |
| ARHGAP15 | 0.0005876 | 1387 | GTEx | DepMap | Descartes | 0.28 | 185.54 |
| MBNL1 | 0.0005242 | 1540 | GTEx | DepMap | Descartes | 0.50 | 152.36 |
| RCSD1 | 0.0005013 | 1612 | GTEx | DepMap | Descartes | 0.06 | 22.93 |
| DOCK10 | 0.0004965 | 1631 | GTEx | DepMap | Descartes | 0.43 | 131.16 |
| ANKRD44 | 0.0004951 | 1634 | GTEx | DepMap | Descartes | 0.36 | 100.58 |
| IKZF1 | 0.0004392 | 1823 | GTEx | DepMap | Descartes | 0.06 | 27.67 |
| SORL1 | 0.0003959 | 2011 | GTEx | DepMap | Descartes | 0.23 | 41.55 |
| WIPF1 | 0.0003886 | 2040 | GTEx | DepMap | Descartes | 0.24 | 99.99 |
| PITPNC1 | 0.0003084 | 2423 | GTEx | DepMap | Descartes | 0.71 | 193.05 |
| SCML4 | 0.0002355 | 2859 | GTEx | DepMap | Descartes | 0.20 | 74.44 |
| NCALD | 0.0001822 | 3291 | GTEx | DepMap | Descartes | 0.25 | 115.10 |
| TOX | 0.0000686 | 4508 | GTEx | DepMap | Descartes | 1.01 | 426.72 |
| EVL | 0.0000181 | 5207 | GTEx | DepMap | Descartes | 0.85 | 422.96 |
| SAMD3 | 0.0000112 | 5316 | GTEx | DepMap | Descartes | 0.05 | 19.33 |
Below shows the ranks on this GEP for immune marker gene lists curated by CellTypist (https://www.celltypist.org/encyclopedia).
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Myelocytes: Myelocytes (model markers)
early granulocyte precursors that are derive from promyelocytes and later mature into metamyelocytes:
Wilcoxon ranksum test P-value for gene set overrepresentation: 8.05e-05
Mean rank of genes in gene set: 162.4
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TYROBP | 0.0076193 | 22 | GTEx | DepMap | Descartes | 0.28 | 1005.51 |
| SRGN | 0.0059010 | 40 | GTEx | DepMap | Descartes | 0.49 | 949.73 |
| S100A6 | 0.0030428 | 129 | GTEx | DepMap | Descartes | 0.54 | 1164.19 |
| FTH1 | 0.0020207 | 273 | GTEx | DepMap | Descartes | 1.44 | 2191.85 |
| ACTB | 0.0017595 | 348 | GTEx | DepMap | Descartes | 3.15 | 2544.66 |
T cells: Treg(diff) (model markers)
unconventional T lymphocyte subpopulation in the thymus which connects αβ T cells and canonical regulatory T cells and is still differentiating:
Wilcoxon ranksum test P-value for gene set overrepresentation: 6.78e-04
Mean rank of genes in gene set: 472.5
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| RGS1 | 0.0062721 | 29 | GTEx | DepMap | Descartes | 0.28 | 263.52 |
| S100A4 | 0.0058178 | 42 | GTEx | DepMap | Descartes | 0.22 | 700.92 |
| PTPRC | 0.0017466 | 350 | GTEx | DepMap | Descartes | 0.19 | 99.90 |
| TSC22D3 | 0.0005528 | 1469 | GTEx | DepMap | Descartes | 0.11 | 70.11 |
Monocytes: Monocytes (model markers)
myeloid mononuclear recirculating leukocytes that are capable of differentiating into macrophages and myeloid lineage dendritic cells:
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.45e-03
Mean rank of genes in gene set: 47
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| NEAT1 | 0.0092469 | 10 | GTEx | DepMap | Descartes | 6.58 | 596.42 |
| TYROBP | 0.0076193 | 22 | GTEx | DepMap | Descartes | 0.28 | 1005.51 |
| SAT1 | 0.0032969 | 109 | GTEx | DepMap | Descartes | 1.33 | 2541.27 |