Program: 22. Neuroblastoma: MYCN.
Submit a comment on this gene expression program’s interpretation: CLICK
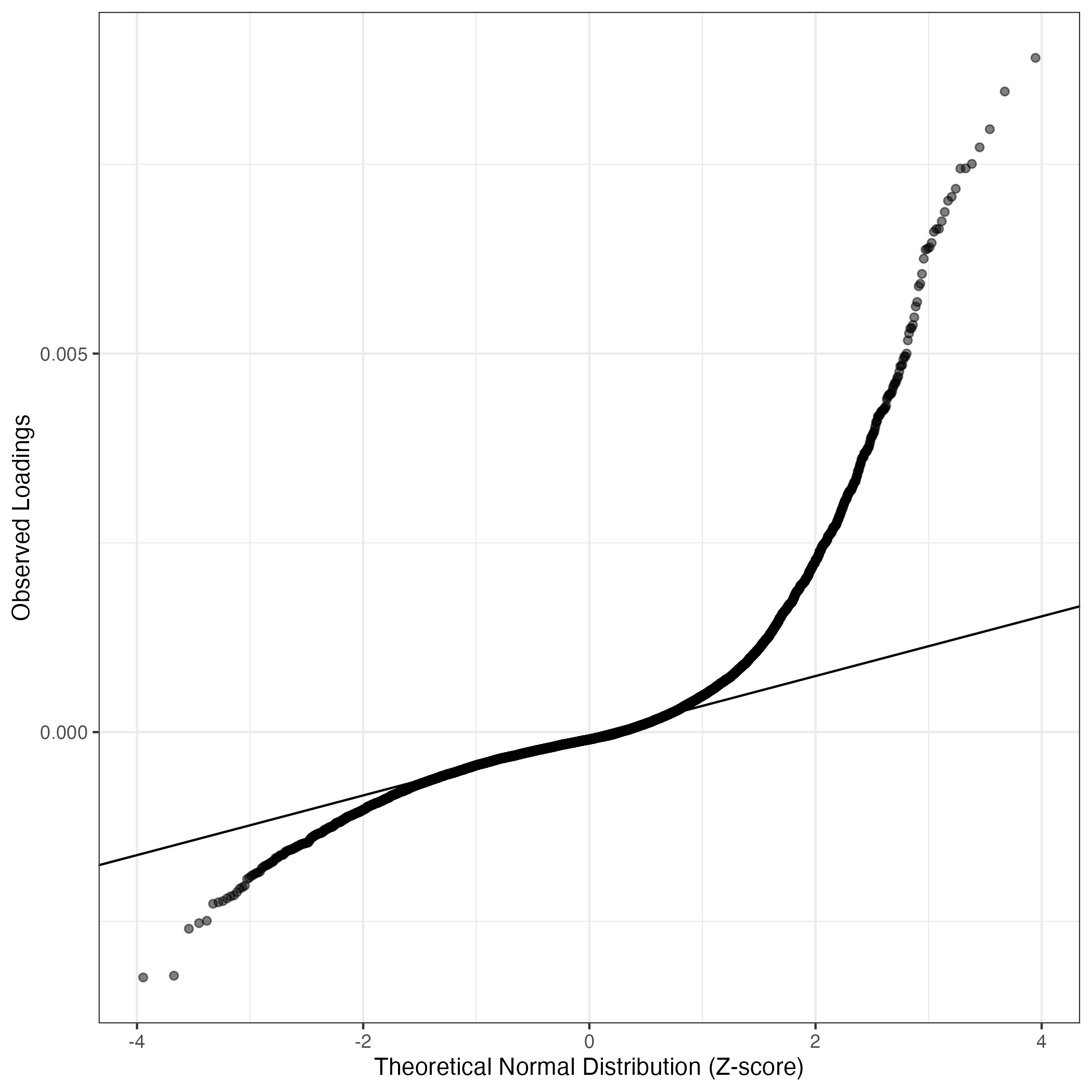
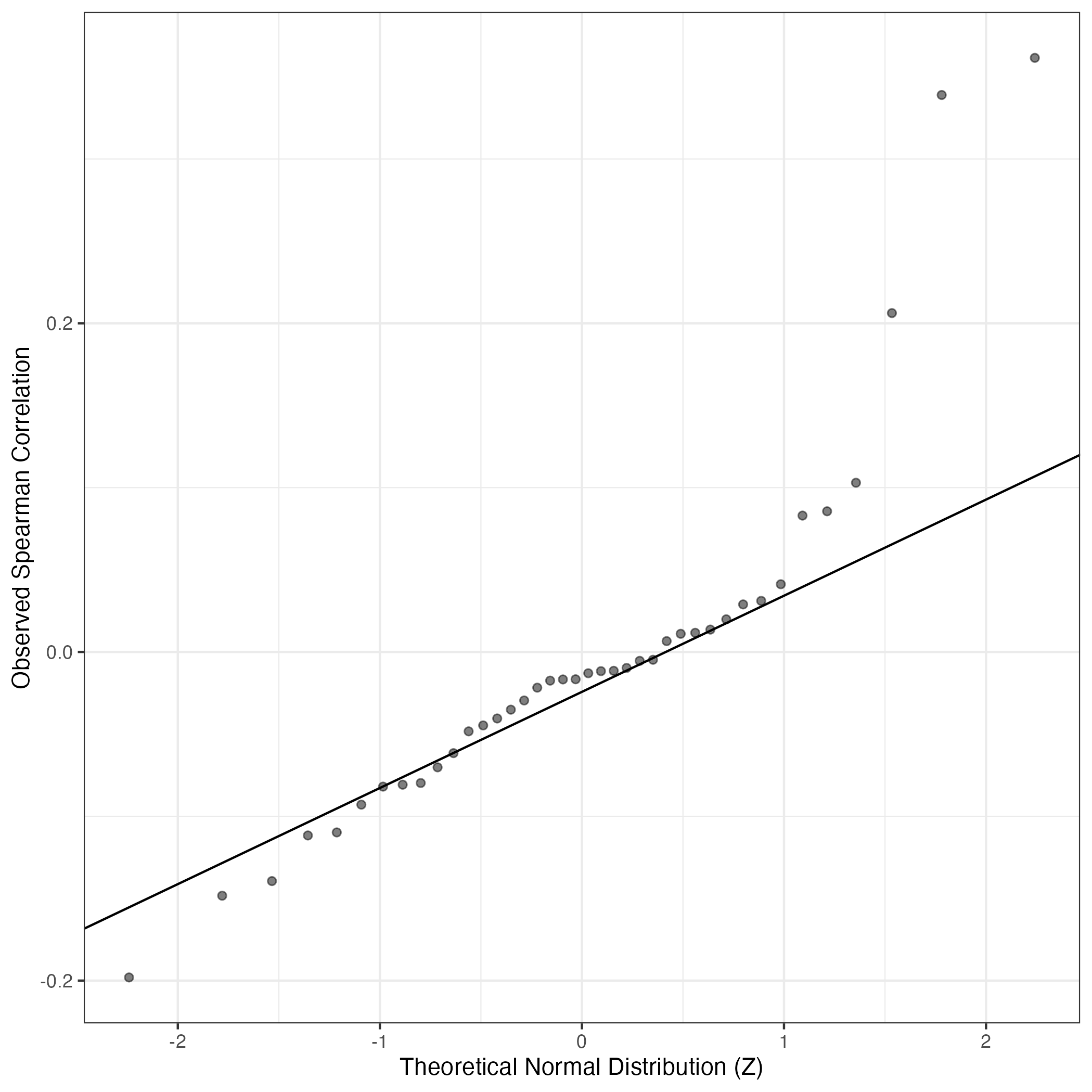
QQ-plot of gene loadings, averaged over both independent splits of the data
This plot highlights the relative contribution of each gene to the GEP
Top genes driving this program.
Note: Decartes website is buggy, try refreshing. Also, Decartes fetal adrenal data have been collected at specific time points (89-122 days), all possible cell types of interest may not be represented, do not overinterpret.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
| Gene | Loading | Gene.Name | GTEx | DepMap | Descartes | Mean.Counts | Mean.Tpm | |
|---|---|---|---|---|---|---|---|---|
| 1 | NPM1 | 0.0089061 | nucleophosmin 1 | GTEx | DepMap | Descartes | 72.58 | 10154.90 |
| 2 | ZFHX4 | 0.0084619 | zinc finger homeobox 4 | GTEx | DepMap | Descartes | 8.61 | 152.50 |
| 3 | EEF1B2 | 0.0079631 | eukaryotic translation elongation factor 1 beta 2 | GTEx | DepMap | Descartes | 68.66 | 14866.95 |
| 4 | EEF2 | 0.0077246 | eukaryotic translation elongation factor 2 | GTEx | DepMap | Descartes | 62.59 | 4741.15 |
| 5 | PEX2 | 0.0075055 | peroxisomal biogenesis factor 2 | GTEx | DepMap | Descartes | 8.60 | 487.15 |
| 6 | NACA | 0.0074465 | nascent polypeptide associated complex subunit alpha | GTEx | DepMap | Descartes | 72.58 | 2600.56 |
| 7 | NPW | 0.0074434 | neuropeptide W | GTEx | DepMap | Descartes | 3.99 | 1265.90 |
| 8 | PKIA | 0.0071768 | cAMP-dependent protein kinase inhibitor alpha | GTEx | DepMap | Descartes | 8.91 | 506.47 |
| 9 | IL7 | 0.0070711 | interleukin 7 | GTEx | DepMap | Descartes | 8.03 | 947.57 |
| 10 | EEF1A1 | 0.0070185 | eukaryotic translation elongation factor 1 alpha 1 | GTEx | DepMap | Descartes | 320.13 | 17070.53 |
| 11 | HNRNPA1 | 0.0068706 | heterogeneous nuclear ribonucleoprotein A1 | GTEx | DepMap | Descartes | 67.17 | 3955.34 |
| 12 | UQCRH | 0.0067470 | ubiquinol-cytochrome c reductase hinge protein | GTEx | DepMap | Descartes | 27.91 | 9104.95 |
| 13 | POLR1D | 0.0066464 | RNA polymerase I and III subunit D | GTEx | DepMap | Descartes | 12.40 | 636.56 |
| 14 | BTF3 | 0.0066455 | basic transcription factor 3 | GTEx | DepMap | Descartes | 51.24 | 9920.10 |
| 15 | TIPIN | 0.0066091 | TIMELESS interacting protein | GTEx | DepMap | Descartes | 14.03 | 1950.25 |
| 16 | ICA1 | 0.0064615 | islet cell autoantigen 1 | GTEx | DepMap | Descartes | 7.34 | 795.28 |
| 17 | EIF3L | 0.0064030 | eukaryotic translation initiation factor 3 subunit L | GTEx | DepMap | Descartes | 23.64 | 1722.74 |
| 18 | ACTN2 | 0.0063845 | actinin alpha 2 | GTEx | DepMap | Descartes | 1.79 | 82.26 |
| 19 | MEIS2 | 0.0063732 | Meis homeobox 2 | GTEx | DepMap | Descartes | 7.19 | 370.74 |
| 20 | CDK6 | 0.0062530 | cyclin dependent kinase 6 | GTEx | DepMap | Descartes | 8.35 | 171.74 |
| 21 | RPSA | 0.0060520 | ribosomal protein SA | GTEx | DepMap | Descartes | 74.56 | 8582.03 |
| 22 | RPL18 | 0.0059225 | ribosomal protein L18 | GTEx | DepMap | Descartes | 98.11 | 5910.89 |
| 23 | ZC2HC1A | 0.0058891 | zinc finger C2HC-type containing 1A | GTEx | DepMap | Descartes | 7.22 | NA |
| 24 | NBEAL1 | 0.0056813 | neurobeachin like 1 | GTEx | DepMap | Descartes | 48.00 | 993.06 |
| 25 | EIF4A2 | 0.0056213 | eukaryotic translation initiation factor 4A2 | GTEx | DepMap | Descartes | 18.87 | 864.39 |
| 26 | GAL | 0.0054785 | galanin and GMAP prepropeptide | GTEx | DepMap | Descartes | 17.53 | 5514.95 |
| 27 | EPCAM | 0.0053776 | epithelial cell adhesion molecule | GTEx | DepMap | Descartes | 1.64 | 241.26 |
| 28 | IFI27 | 0.0053356 | interferon alpha inducible protein 27 | GTEx | DepMap | Descartes | 19.27 | 1979.87 |
| 29 | ARL9 | 0.0053330 | ADP ribosylation factor like GTPase 9 | GTEx | DepMap | Descartes | 0.57 | 90.09 |
| 30 | MYC | 0.0052663 | MYC proto-oncogene, bHLH transcription factor | GTEx | DepMap | Descartes | 9.01 | 600.21 |
| 31 | MYL6B | 0.0051744 | myosin light chain 6B | GTEx | DepMap | Descartes | 6.67 | 1180.64 |
| 32 | GLCCI1 | 0.0050009 | glucocorticoid induced 1 | GTEx | DepMap | Descartes | 5.04 | 261.82 |
| 33 | CCNI | 0.0049622 | cyclin I | GTEx | DepMap | Descartes | 22.13 | 1804.42 |
| 34 | PHB2 | 0.0049616 | prohibitin 2 | GTEx | DepMap | Descartes | 6.97 | 1099.10 |
| 35 | PPP1R14B | 0.0049223 | protein phosphatase 1 regulatory inhibitor subunit 14B | GTEx | DepMap | Descartes | 7.15 | 1441.12 |
| 36 | PHOX2A | 0.0048503 | paired like homeobox 2A | GTEx | DepMap | Descartes | 7.94 | 1160.99 |
| 37 | TTC3 | 0.0048378 | tetratricopeptide repeat domain 3 | GTEx | DepMap | Descartes | 12.11 | NA |
| 38 | EBPL | 0.0048313 | EBP like | GTEx | DepMap | Descartes | 4.48 | 1048.31 |
| 39 | RPL13 | 0.0047588 | ribosomal protein L13 | GTEx | DepMap | Descartes | 247.80 | 12732.50 |
| 40 | YBX1 | 0.0046965 | Y-box binding protein 1 | GTEx | DepMap | Descartes | 46.08 | 3316.69 |
| 41 | EIF3E | 0.0046828 | eukaryotic translation initiation factor 3 subunit E | GTEx | DepMap | Descartes | 18.31 | 2123.68 |
| 42 | NME2 | 0.0046404 | NME/NM23 nucleoside diphosphate kinase 2 | GTEx | DepMap | Descartes | 5.13 | 1232.97 |
| 43 | AHCY | 0.0046103 | adenosylhomocysteinase | GTEx | DepMap | Descartes | 3.78 | 344.92 |
| 44 | DLL3 | 0.0046056 | delta like canonical Notch ligand 3 | GTEx | DepMap | Descartes | 0.98 | 93.22 |
| 45 | MDK | 0.0045787 | midkine | GTEx | DepMap | Descartes | 10.30 | 1971.22 |
| 46 | MEG3 | 0.0045510 | maternally expressed 3 | GTEx | DepMap | Descartes | 58.05 | 1267.76 |
| 47 | KHDRBS1 | 0.0045028 | KH RNA binding domain containing, signal transduction associated 1 | GTEx | DepMap | Descartes | 9.10 | 764.78 |
| 48 | MUC4 | 0.0044736 | mucin 4, cell surface associated | GTEx | DepMap | Descartes | 0.54 | 6.09 |
| 49 | NSA2 | 0.0044644 | NSA2 ribosome biogenesis factor | GTEx | DepMap | Descartes | 7.35 | 394.25 |
| 50 | RSPO4 | 0.0044594 | R-spondin 4 | GTEx | DepMap | Descartes | 0.51 | 43.51 |
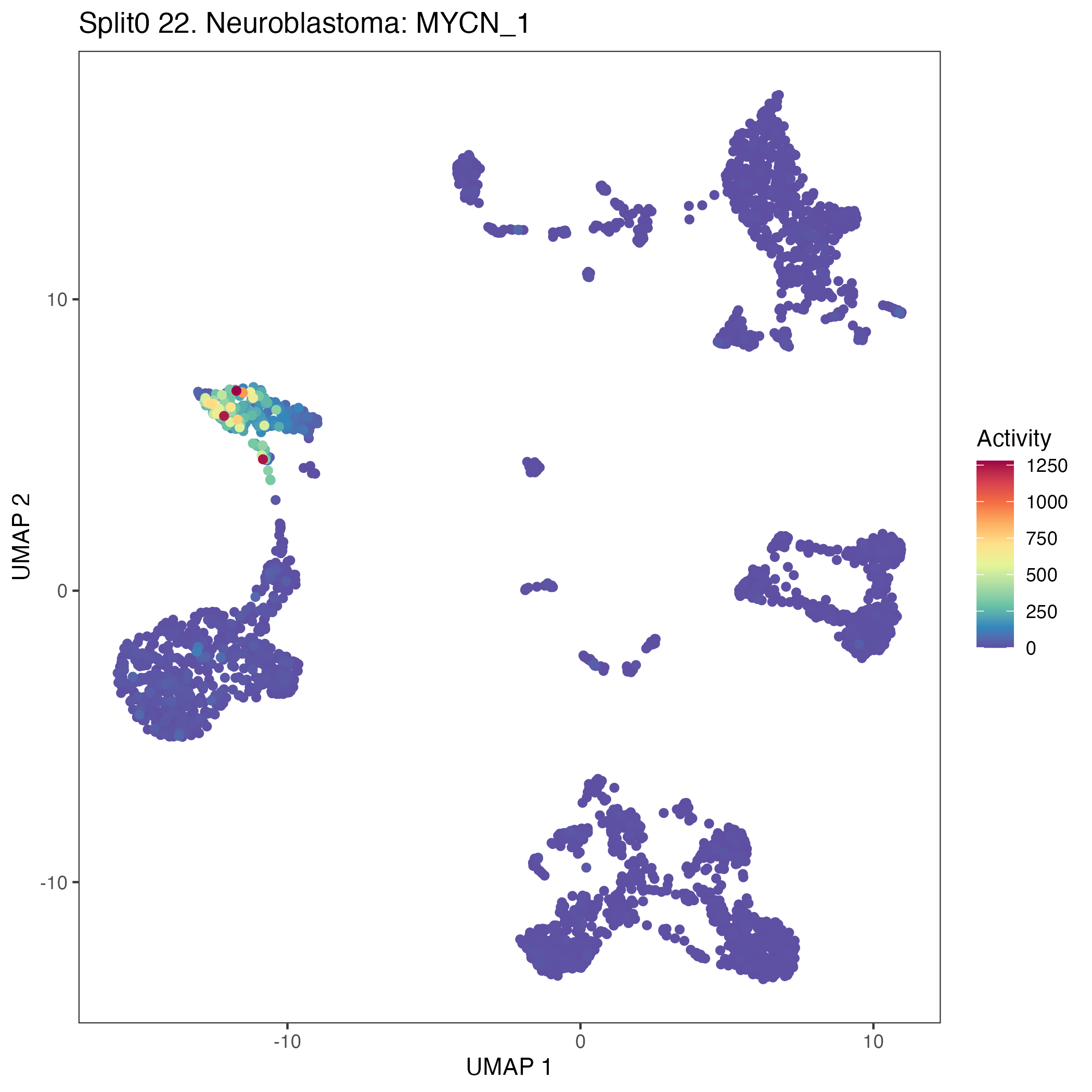
UMAP plots showing activity of gene expression program identified in GEP 22. Neuroblastoma: MYCN:
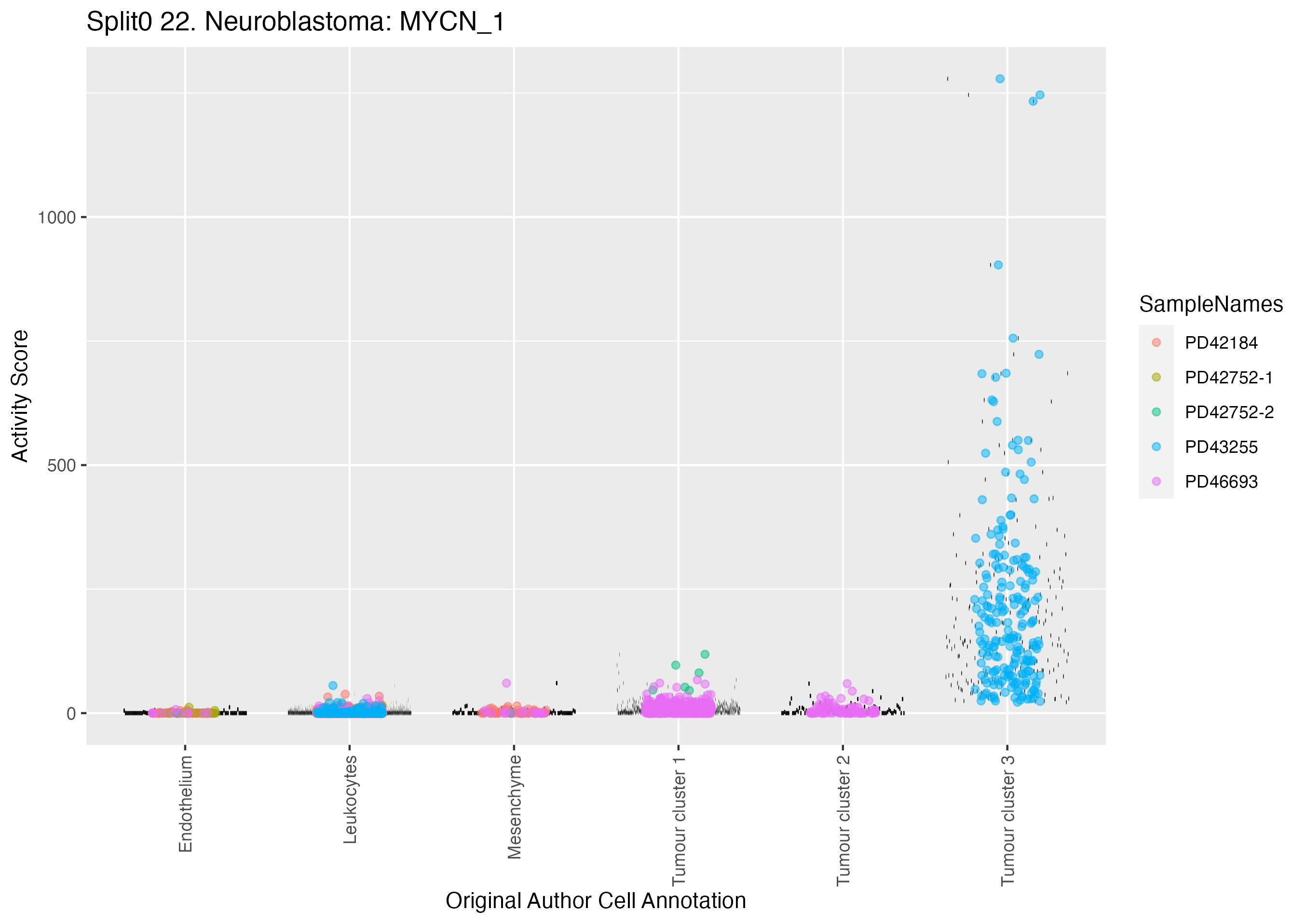
Boxlot showing activity of gene expression program identified in GEP 22. Neuroblastoma: MYCN:
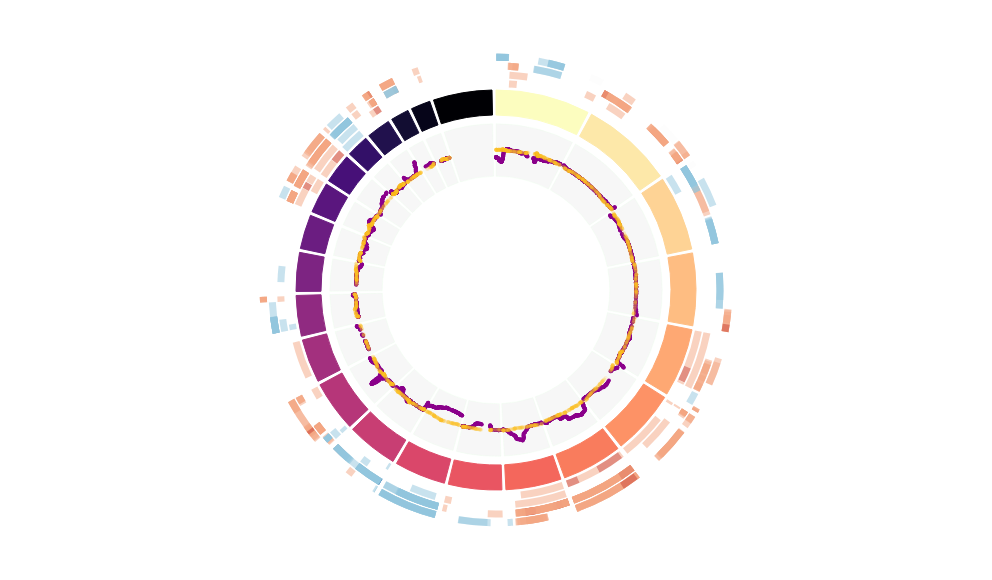
CNV Data procured from inferCNV.
Outer tracks are putative CNV regions (gains = red, losses = blue) for each patient
Inner track is expression data representing:
The top cells expressing this GEP (purple)
Random cells (n =50) from the reference set used in inferCNV (orange)
Gene set Enrichments for this program, caculated from top 50 genes
mSigDB Cell Types Gene Set:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| RUBENSTEIN_SKELETAL_MUSCLE_SATELLITE_CELLS | 4.16e-17 | 24.08 | 12.57 | 9.84e-15 | 2.79e-14 | 18NPM1, EEF1B2, EEF2, NACA, EEF1A1, HNRNPA1, UQCRH, BTF3, EIF3L, RPSA, RPL18, EIF4A2, MYC, PPP1R14B, TTC3, RPL13, EIF3E, MEG3 |
310 |
| BUSSLINGER_DUODENAL_STEM_CELLS | 4.40e-17 | 24.00 | 12.53 | 9.84e-15 | 2.95e-14 | 18NPM1, EEF1B2, EEF2, EEF1A1, HNRNPA1, BTF3, EIF3L, CDK6, RPL18, EIF4A2, EPCAM, TTC3, RPL13, YBX1, EIF3E, AHCY, KHDRBS1, NSA2 |
311 |
| FAN_EMBRYONIC_CTX_EX_2_EXCITATORY_NEURON | 3.20e-05 | 61.17 | 10.83 | 8.59e-04 | 2.15e-02 | 3ZFHX4, MEIS2, MDK |
16 |
| TRAVAGLINI_LUNG_PROXIMAL_BASAL_CELL | 8.88e-18 | 17.59 | 9.57 | 5.96e-15 | 5.96e-15 | 23NPM1, EEF1B2, EEF2, NACA, EEF1A1, HNRNPA1, UQCRH, POLR1D, BTF3, EIF3L, MEIS2, RPSA, RPL18, EIF4A2, MYL6B, CCNI, PHB2, EBPL, RPL13, YBX1, EIF3E, AHCY, MDK |
629 |
| BUSSLINGER_DUODENAL_TRANSIT_AMPLIFYING_CELLS | 1.71e-10 | 19.26 | 8.76 | 1.64e-08 | 1.15e-07 | 11NPM1, EEF1B2, EEF2, EEF1A1, HNRNPA1, EIF3L, RPL18, EPCAM, PHB2, RPL13, YBX1 |
194 |
| FAN_OVARY_CL0_XBP1_SELK_HIGH_STROMAL_CELL | 3.84e-09 | 20.46 | 8.57 | 2.34e-07 | 2.58e-06 | 9NPM1, NACA, BTF3, RPL18, EIF4A2, RPL13, YBX1, EIF3E, MEG3 |
143 |
| BUSSLINGER_GASTRIC_PPP1R1B_POSITIVE_CELLS | 2.10e-08 | 21.07 | 8.35 | 1.09e-06 | 1.41e-05 | 8NPM1, EEF1B2, EEF2, EEF1A1, HNRNPA1, RPL18, EPCAM, RPL13 |
121 |
| BUSSLINGER_DUODENAL_DIFFERENTIATING_STEM_CELLS | 5.80e-12 | 16.71 | 8.23 | 7.78e-10 | 3.89e-09 | 14NPM1, EEF1B2, EEF2, EEF1A1, HNRNPA1, BTF3, EIF3L, RPL18, EPCAM, PHB2, RPL13, YBX1, EIF3E, AHCY |
305 |
| RUBENSTEIN_SKELETAL_MUSCLE_T_CELLS | 1.65e-09 | 18.27 | 8.01 | 1.23e-07 | 1.11e-06 | 10NPM1, EEF1B2, EEF1A1, HNRNPA1, BTF3, EIF3L, RPSA, RPL18, RPL13, EIF3E |
181 |
| LAKE_ADULT_KIDNEY_C8_DECENDING_THIN_LIMB | 3.05e-11 | 16.58 | 7.99 | 3.41e-09 | 2.05e-08 | 13NPM1, EEF1B2, EEF2, NACA, EEF1A1, BTF3, RPSA, RPL18, EIF4A2, EPCAM, CCNI, RPL13, YBX1 |
278 |
| LAKE_ADULT_KIDNEY_C9_THIN_ASCENDING_LIMB | 3.27e-10 | 15.42 | 7.25 | 2.75e-08 | 2.20e-07 | 12NPM1, EEF2, NACA, EEF1A1, BTF3, RPSA, RPL18, EIF4A2, EPCAM, CCNI, RPL13, YBX1 |
268 |
| TRAVAGLINI_LUNG_BASAL_CELL | 4.08e-08 | 15.33 | 6.45 | 1.96e-06 | 2.74e-05 | 9NPM1, NACA, EEF1A1, HNRNPA1, BTF3, RPSA, MYC, RPL13, AHCY |
188 |
| BUSSLINGER_ESOPHAGEAL_PROLIFERATING_BASAL_CELLS | 7.93e-06 | 21.34 | 6.41 | 2.53e-04 | 5.32e-03 | 5EEF1B2, EEF2, EEF1A1, HNRNPA1, YBX1 |
70 |
| LAKE_ADULT_KIDNEY_C18_COLLECTING_DUCT_PRINCIPAL_CELLS_MEDULLA | 2.54e-09 | 12.74 | 6.00 | 1.70e-07 | 1.70e-06 | 12NPM1, EEF2, NACA, EEF1A1, BTF3, RPSA, RPL18, EIF4A2, EPCAM, CCNI, RPL13, YBX1 |
322 |
| LAKE_ADULT_KIDNEY_C7_PROXIMAL_TUBULE_EPITHELIAL_CELLS_S3 | 9.03e-07 | 15.89 | 5.92 | 3.19e-05 | 6.06e-04 | 7NPM1, EEF2, EEF1A1, RPSA, RPL18, CCNI, RPL13 |
135 |
| TRAVAGLINI_LUNG_PROLIFERATING_BASAL_CELL | 1.51e-12 | 10.42 | 5.62 | 2.53e-10 | 1.01e-09 | 21NPM1, EEF1B2, EEF2, NACA, EEF1A1, HNRNPA1, UQCRH, POLR1D, BTF3, EIF3L, RPSA, RPL18, MYL6B, PHB2, PPP1R14B, RPL13, YBX1, EIF3E, AHCY, MDK, KHDRBS1 |
891 |
| TRAVAGLINI_LUNG_BRONCHIAL_VESSEL_1_CELL | 1.79e-07 | 12.77 | 5.39 | 8.02e-06 | 1.20e-04 | 9NPM1, EEF2, NACA, HNRNPA1, RPSA, RPL18, MYC, RPL13, EIF3E |
224 |
| FAN_OVARY_CL1_GPRC5A_TNFRS12A_HIGH_SELECTABLE_FOLLICLE_STROMAL_CELL | 1.55e-08 | 10.73 | 5.06 | 8.66e-07 | 1.04e-05 | 12NPM1, EEF2, NACA, EEF1A1, HNRNPA1, RPSA, RPL18, MYC, CCNI, YBX1, EIF3E, MEG3 |
380 |
| MENON_FETAL_KIDNEY_1_EMBRYONIC_RED_BLOOD_CELLS | 9.51e-06 | 14.20 | 4.85 | 2.90e-04 | 6.38e-03 | 6NPM1, EEF1B2, NACA, BTF3, RPSA, YBX1 |
126 |
| HAY_BONE_MARROW_EARLY_ERYTHROBLAST | 3.49e-05 | 15.42 | 4.67 | 9.01e-04 | 2.34e-02 | 5NPM1, UQCRH, GAL, MYC, AHCY |
95 |
Dowload full table
mSigDB Hallmark Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| HALLMARK_MYC_TARGETS_V1 | 1.24e-04 | 8.78 | 3.02 | 6.18e-03 | 6.18e-03 | 6NPM1, EEF1B2, HNRNPA1, RPL18, MYC, PHB2 |
200 |
| HALLMARK_MYOGENESIS | 7.71e-03 | 5.55 | 1.44 | 1.67e-01 | 3.85e-01 | 4PKIA, ACTN2, EIF4A2, MYL6B |
200 |
| HALLMARK_UNFOLDED_PROTEIN_RESPONSE | 1.00e-02 | 7.25 | 1.42 | 1.67e-01 | 5.00e-01 | 3NPM1, EEF2, EIF4A2 |
113 |
| HALLMARK_MYC_TARGETS_V2 | 2.21e-02 | 9.30 | 1.07 | 2.77e-01 | 1.00e+00 | 2NPM1, MYC |
58 |
| HALLMARK_INTERFERON_ALPHA_RESPONSE | 5.62e-02 | 5.48 | 0.64 | 5.44e-01 | 1.00e+00 | 2IL7, IFI27 |
97 |
| HALLMARK_UV_RESPONSE_DN | 1.10e-01 | 3.67 | 0.43 | 5.44e-01 | 1.00e+00 | 2ICA1, MYC |
144 |
| HALLMARK_UV_RESPONSE_UP | 1.28e-01 | 3.34 | 0.39 | 5.44e-01 | 1.00e+00 | 2GAL, EPCAM |
158 |
| HALLMARK_IL2_STAT5_SIGNALING | 1.83e-01 | 2.64 | 0.31 | 5.44e-01 | 1.00e+00 | 2MYC, AHCY |
199 |
| HALLMARK_G2M_CHECKPOINT | 1.85e-01 | 2.63 | 0.31 | 5.44e-01 | 1.00e+00 | 2MEIS2, MYC |
200 |
| HALLMARK_ESTROGEN_RESPONSE_LATE | 1.85e-01 | 2.63 | 0.31 | 5.44e-01 | 1.00e+00 | 2GAL, MDK |
200 |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | 1.85e-01 | 2.63 | 0.31 | 5.44e-01 | 1.00e+00 | 2IL7, IFI27 |
200 |
| HALLMARK_APICAL_JUNCTION | 1.85e-01 | 2.63 | 0.31 | 5.44e-01 | 1.00e+00 | 2ACTN2, MDK |
200 |
| HALLMARK_E2F_TARGETS | 1.85e-01 | 2.63 | 0.31 | 5.44e-01 | 1.00e+00 | 2TIPIN, MYC |
200 |
| HALLMARK_OXIDATIVE_PHOSPHORYLATION | 1.85e-01 | 2.63 | 0.31 | 5.44e-01 | 1.00e+00 | 2UQCRH, PHB2 |
200 |
| HALLMARK_ALLOGRAFT_REJECTION | 1.85e-01 | 2.63 | 0.31 | 5.44e-01 | 1.00e+00 | 2NPM1, IL7 |
200 |
| HALLMARK_HEDGEHOG_SIGNALING | 1.34e-01 | 7.29 | 0.18 | 5.44e-01 | 1.00e+00 | 1CDK6 |
36 |
| HALLMARK_WNT_BETA_CATENIN_SIGNALING | 1.54e-01 | 6.22 | 0.15 | 5.44e-01 | 1.00e+00 | 1MYC |
42 |
| HALLMARK_IL6_JAK_STAT3_SIGNALING | 2.93e-01 | 2.97 | 0.07 | 8.06e-01 | 1.00e+00 | 1IL7 |
87 |
| HALLMARK_PROTEIN_SECRETION | 3.17e-01 | 2.68 | 0.07 | 8.06e-01 | 1.00e+00 | 1ICA1 |
96 |
| HALLMARK_ANDROGEN_RESPONSE | 3.28e-01 | 2.58 | 0.06 | 8.06e-01 | 1.00e+00 | 1CDK6 |
100 |
Dowload full table
KEGG Pathways:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| KEGG_RIBOSOME | 5.05e-03 | 9.38 | 1.83 | 9.39e-01 | 9.39e-01 | 3RPSA, RPL18, RPL13 |
88 |
| KEGG_CHRONIC_MYELOID_LEUKEMIA | 3.38e-02 | 7.33 | 0.85 | 1.00e+00 | 1.00e+00 | 2CDK6, MYC |
73 |
| KEGG_SMALL_CELL_LUNG_CANCER | 4.36e-02 | 6.35 | 0.74 | 1.00e+00 | 1.00e+00 | 2CDK6, MYC |
84 |
| KEGG_PYRIMIDINE_METABOLISM | 5.73e-02 | 5.42 | 0.63 | 1.00e+00 | 1.00e+00 | 2POLR1D, NME2 |
98 |
| KEGG_CELL_CYCLE | 8.71e-02 | 4.23 | 0.49 | 1.00e+00 | 1.00e+00 | 2CDK6, MYC |
125 |
| KEGG_JAK_STAT_SIGNALING_PATHWAY | 1.24e-01 | 3.40 | 0.40 | 1.00e+00 | 1.00e+00 | 2IL7, MYC |
155 |
| KEGG_PURINE_METABOLISM | 1.29e-01 | 3.32 | 0.39 | 1.00e+00 | 1.00e+00 | 2POLR1D, NME2 |
159 |
| KEGG_SELENOAMINO_ACID_METABOLISM | 9.85e-02 | 10.19 | 0.24 | 1.00e+00 | 1.00e+00 | 1AHCY |
26 |
| KEGG_RNA_POLYMERASE | 1.09e-01 | 9.11 | 0.22 | 1.00e+00 | 1.00e+00 | 1POLR1D |
29 |
| KEGG_THYROID_CANCER | 1.09e-01 | 9.11 | 0.22 | 1.00e+00 | 1.00e+00 | 1MYC |
29 |
| KEGG_PATHWAYS_IN_CANCER | 3.61e-01 | 1.61 | 0.19 | 1.00e+00 | 1.00e+00 | 2CDK6, MYC |
325 |
| KEGG_CYSTEINE_AND_METHIONINE_METABOLISM | 1.27e-01 | 7.73 | 0.19 | 1.00e+00 | 1.00e+00 | 1AHCY |
34 |
| KEGG_BLADDER_CANCER | 1.54e-01 | 6.22 | 0.15 | 1.00e+00 | 1.00e+00 | 1MYC |
42 |
| KEGG_TYPE_I_DIABETES_MELLITUS | 1.57e-01 | 6.07 | 0.15 | 1.00e+00 | 1.00e+00 | 1ICA1 |
43 |
| KEGG_NOTCH_SIGNALING_PATHWAY | 1.71e-01 | 5.54 | 0.13 | 1.00e+00 | 1.00e+00 | 1DLL3 |
47 |
| KEGG_ENDOMETRIAL_CANCER | 1.87e-01 | 5.00 | 0.12 | 1.00e+00 | 1.00e+00 | 1MYC |
52 |
| KEGG_NON_SMALL_CELL_LUNG_CANCER | 1.94e-01 | 4.81 | 0.12 | 1.00e+00 | 1.00e+00 | 1CDK6 |
54 |
| KEGG_CYTOSOLIC_DNA_SENSING_PATHWAY | 1.97e-01 | 4.72 | 0.12 | 1.00e+00 | 1.00e+00 | 1POLR1D |
55 |
| KEGG_ACUTE_MYELOID_LEUKEMIA | 2.03e-01 | 4.55 | 0.11 | 1.00e+00 | 1.00e+00 | 1MYC |
57 |
| KEGG_COLORECTAL_CANCER | 2.19e-01 | 4.18 | 0.10 | 1.00e+00 | 1.00e+00 | 1MYC |
62 |
Dowload full table
CHR Positional Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| chr8q21 | 6.44e-04 | 8.03 | 2.46 | 1.79e-01 | 1.79e-01 | 5ZFHX4, PEX2, PKIA, IL7, ZC2HC1A |
178 |
| chr7p21 | 4.26e-02 | 6.43 | 0.74 | 1.00e+00 | 1.00e+00 | 2ICA1, GLCCI1 |
83 |
| chr5q13 | 1.08e-01 | 3.72 | 0.43 | 1.00e+00 | 1.00e+00 | 2BTF3, NSA2 |
142 |
| chr12q13 | 2.07e-01 | 1.98 | 0.39 | 1.00e+00 | 1.00e+00 | 3NACA, HNRNPA1, MYL6B |
407 |
| chr11q13 | 2.21e-01 | 1.91 | 0.38 | 1.00e+00 | 1.00e+00 | 3GAL, PPP1R14B, PHOX2A |
421 |
| chr2q33 | 1.78e-01 | 2.70 | 0.32 | 1.00e+00 | 1.00e+00 | 2EEF1B2, NBEAL1 |
195 |
| chr8q23 | 1.61e-01 | 5.93 | 0.14 | 1.00e+00 | 1.00e+00 | 1EIF3E |
44 |
| chr15q14 | 2.00e-01 | 4.64 | 0.11 | 1.00e+00 | 1.00e+00 | 1MEIS2 |
56 |
| chr6q13 | 2.00e-01 | 4.64 | 0.11 | 1.00e+00 | 1.00e+00 | 1EEF1A1 |
56 |
| chr14q32 | 1.00e+00 | 0.96 | 0.11 | 1.00e+00 | 1.00e+00 | 2IFI27, MEG3 |
546 |
| chr1p33 | 2.13e-01 | 4.32 | 0.11 | 1.00e+00 | 1.00e+00 | 1UQCRH |
60 |
| chr1q43 | 2.13e-01 | 4.32 | 0.11 | 1.00e+00 | 1.00e+00 | 1ACTN2 |
60 |
| chr4q12 | 2.70e-01 | 3.27 | 0.08 | 1.00e+00 | 1.00e+00 | 1ARL9 |
79 |
| chr2p21 | 2.73e-01 | 3.23 | 0.08 | 1.00e+00 | 1.00e+00 | 1EPCAM |
80 |
| chr3q27 | 3.67e-01 | 2.24 | 0.06 | 1.00e+00 | 1.00e+00 | 1EIF4A2 |
115 |
| chr20p13 | 3.72e-01 | 2.20 | 0.05 | 1.00e+00 | 1.00e+00 | 1RSPO4 |
117 |
| chr19q13 | 4.39e-01 | 0.45 | 0.05 | 1.00e+00 | 1.00e+00 | 2RPL18, DLL3 |
1165 |
| chr3q29 | 3.84e-01 | 2.11 | 0.05 | 1.00e+00 | 1.00e+00 | 1MUC4 |
122 |
| chr15q22 | 3.89e-01 | 2.07 | 0.05 | 1.00e+00 | 1.00e+00 | 1TIPIN |
124 |
| chr1p35 | 4.03e-01 | 1.98 | 0.05 | 1.00e+00 | 1.00e+00 | 1KHDRBS1 |
130 |
Dowload full table
Transcription Factor Targets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GTF2A2_TARGET_GENES | 2.30e-05 | 6.11 | 2.71 | 2.61e-02 | 2.61e-02 | 10NPM1, EEF2, EEF1A1, HNRNPA1, RPSA, RPL18, EIF4A2, MYC, CCNI, RPL13 |
522 |
| PSMB5_TARGET_GENES | 1.16e-03 | 5.66 | 1.96 | 3.99e-01 | 1.00e+00 | 6NPM1, EEF2, EEF1A1, RPSA, EIF4A2, PPP1R14B |
307 |
| PER1_TARGET_GENES | 7.55e-03 | 16.78 | 1.89 | 7.81e-01 | 1.00e+00 | 2EEF1A1, EIF4A2 |
33 |
| GCCATNTTG_YY1_Q6 | 1.33e-03 | 4.73 | 1.79 | 3.99e-01 | 1.00e+00 | 7EEF1B2, NACA, HNRNPA1, UQCRH, BTF3, PHB2, YBX1 |
437 |
| YY1_Q6 | 2.88e-03 | 5.65 | 1.73 | 6.51e-01 | 1.00e+00 | 5NACA, EEF1A1, UQCRH, BTF3, YBX1 |
251 |
| FOXE1_TARGET_GENES | 1.41e-03 | 3.82 | 1.63 | 3.99e-01 | 1.00e+00 | 9EEF2, NACA, PKIA, HNRNPA1, UQCRH, RPL18, EIF4A2, RPL13, YBX1 |
728 |
| RUVBL1_TARGET_GENES | 5.37e-03 | 6.17 | 1.60 | 7.26e-01 | 1.00e+00 | 4NACA, EEF1A1, RPL18, PHB2 |
180 |
| ADA2_TARGET_GENES | 3.69e-03 | 3.28 | 1.40 | 6.97e-01 | 1.00e+00 | 9ZFHX4, EEF2, EEF1A1, MEIS2, RPL18, EIF4A2, PHB2, RPL13, NSA2 |
846 |
| ATF5_TARGET_GENES | 7.89e-03 | 3.77 | 1.31 | 7.81e-01 | 1.00e+00 | 6UQCRH, BTF3, MEIS2, EIF4A2, EPCAM, NSA2 |
458 |
| KAT5_TARGET_GENES | 5.77e-03 | 3.05 | 1.30 | 7.26e-01 | 1.00e+00 | 9EEF1B2, NACA, NPW, EEF1A1, UQCRH, CDK6, RPSA, RPL13, EIF3E |
910 |
| PHF2_TARGET_GENES | 5.02e-03 | 2.68 | 1.27 | 7.26e-01 | 1.00e+00 | 12NACA, HNRNPA1, POLR1D, EIF3L, MEIS2, EIF4A2, CCNI, PPP1R14B, RPL13, AHCY, KHDRBS1, NSA2 |
1485 |
| EMX1_TARGET_GENES | 1.16e-02 | 4.90 | 1.27 | 8.35e-01 | 1.00e+00 | 4NPM1, UQCRH, EIF4A2, CCNI |
226 |
| PBX1_02 | 1.51e-02 | 6.18 | 1.22 | 8.35e-01 | 1.00e+00 | 3IL7, MEIS2, KHDRBS1 |
132 |
| E2F_Q3_01 | 1.34e-02 | 4.69 | 1.21 | 8.35e-01 | 1.00e+00 | 4HNRNPA1, TIPIN, MEIS2, MYC |
236 |
| E2F_Q4_01 | 1.38e-02 | 4.65 | 1.20 | 8.35e-01 | 1.00e+00 | 4HNRNPA1, TIPIN, MEIS2, MYC |
238 |
| PR_02 | 1.61e-02 | 6.04 | 1.19 | 8.35e-01 | 1.00e+00 | 3ZFHX4, EEF1B2, KHDRBS1 |
135 |
| E2F1_Q6_01 | 1.48e-02 | 4.55 | 1.18 | 8.35e-01 | 1.00e+00 | 4HNRNPA1, MEIS2, RPL18, MYC |
243 |
| SMAD4_Q6 | 1.50e-02 | 4.53 | 1.17 | 8.35e-01 | 1.00e+00 | 4EEF1A1, MEIS2, CDK6, MYL6B |
244 |
| SNRNP70_TARGET_GENES | 1.06e-02 | 2.75 | 1.17 | 8.35e-01 | 1.00e+00 | 9EEF2, NACA, EEF1A1, HNRNPA1, RPL18, EIF4A2, RPL13, NME2, AHCY |
1009 |
| NFMUE1_Q6 | 1.62e-02 | 4.42 | 1.15 | 8.35e-01 | 1.00e+00 | 4NACA, EEF1A1, HNRNPA1, UQCRH |
250 |
Dowload full table
GO Biological Processes:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GOBP_NEGATIVE_REGULATION_OF_MONOCYTE_DIFFERENTIATION | 4.28e-04 | 86.37 | 8.33 | 3.89e-01 | 1.00e+00 | 2CDK6, MYC |
8 |
| GOBP_CYTOPLASMIC_TRANSLATION | 2.82e-06 | 17.74 | 6.04 | 1.06e-02 | 2.11e-02 | 6EEF2, EIF3L, RPSA, RPL18, EIF4A2, EIF3E |
102 |
| GOBP_TRANSLATIONAL_INITIATION | 7.46e-07 | 12.87 | 5.15 | 5.58e-03 | 5.58e-03 | 8NPM1, EIF3L, RPSA, RPL18, EIF4A2, RPL13, EIF3E, KHDRBS1 |
193 |
| GOBP_CYTOPLASMIC_TRANSLATIONAL_INITIATION | 3.24e-04 | 25.71 | 4.86 | 3.46e-01 | 1.00e+00 | 3EIF3L, EIF4A2, EIF3E |
34 |
| GOBP_FORMATION_OF_CYTOPLASMIC_TRANSLATION_INITIATION_COMPLEX | 1.80e-03 | 37.11 | 3.99 | 9.14e-01 | 1.00e+00 | 2EIF3L, EIF3E |
16 |
| GOBP_REGULATION_OF_TRANSLATIONAL_INITIATION | 2.76e-04 | 14.30 | 3.65 | 3.44e-01 | 1.00e+00 | 4NPM1, EIF4A2, EIF3E, KHDRBS1 |
80 |
| GOBP_NEGATIVE_REGULATION_OF_MYELOID_LEUKOCYTE_DIFFERENTIATION | 8.97e-04 | 17.71 | 3.40 | 5.60e-01 | 1.00e+00 | 3CDK6, MYC, NME2 |
48 |
| GOBP_NEGATIVE_REGULATION_OF_MYELOID_CELL_DIFFERENTIATION | 4.68e-04 | 12.34 | 3.16 | 3.89e-01 | 1.00e+00 | 4MEIS2, CDK6, MYC, NME2 |
92 |
| GOBP_REGULATION_OF_MONOCYTE_DIFFERENTIATION | 3.10e-03 | 27.37 | 3.01 | 1.00e+00 | 1.00e+00 | 2CDK6, MYC |
21 |
| GOBP_NEGATIVE_REGULATION_OF_CELLULAR_SENESCENCE | 3.40e-03 | 26.01 | 2.87 | 1.00e+00 | 1.00e+00 | 2CDK6, YBX1 |
22 |
| GOBP_NEGATIVE_REGULATION_OF_HEMOPOIESIS | 7.69e-04 | 10.75 | 2.76 | 5.60e-01 | 1.00e+00 | 4CDK6, MYC, NME2, MDK |
105 |
| GOBP_PEPTIDE_BIOSYNTHETIC_PROCESS | 1.39e-05 | 5.43 | 2.57 | 3.47e-02 | 1.04e-01 | 12NPM1, EEF1B2, EEF2, EEF1A1, EIF3L, RPSA, RPL18, EIF4A2, RPL13, YBX1, EIF3E, KHDRBS1 |
740 |
| GOBP_POSITIVE_REGULATION_OF_POTASSIUM_ION_TRANSMEMBRANE_TRANSPORTER_ACTIVITY | 4.39e-03 | 22.60 | 2.52 | 1.00e+00 | 1.00e+00 | 2ACTN2, GAL |
25 |
| GOBP_NUCLEAR_TRANSCRIBED_MRNA_CATABOLIC_PROCESS_NONSENSE_MEDIATED_DECAY | 1.26e-03 | 9.37 | 2.41 | 7.25e-01 | 1.00e+00 | 4RPSA, RPL18, RPL13, EIF3E |
120 |
| GOBP_NEGATIVE_REGULATION_OF_CELL_AGING | 5.48e-03 | 20.00 | 2.24 | 1.00e+00 | 1.00e+00 | 2CDK6, YBX1 |
28 |
| GOBP_POSITIVE_REGULATION_OF_TRANSLATION | 1.83e-03 | 8.43 | 2.17 | 9.14e-01 | 1.00e+00 | 4NPM1, EEF2, EIF3E, KHDRBS1 |
133 |
| GOBP_AMIDE_BIOSYNTHETIC_PROCESS | 6.73e-05 | 4.57 | 2.16 | 1.26e-01 | 5.04e-01 | 12NPM1, EEF1B2, EEF2, EEF1A1, EIF3L, RPSA, RPL18, EIF4A2, RPL13, YBX1, EIF3E, KHDRBS1 |
877 |
| GOBP_PEPTIDE_METABOLIC_PROCESS | 8.76e-05 | 4.43 | 2.10 | 1.31e-01 | 6.55e-01 | 12NPM1, EEF1B2, EEF2, EEF1A1, EIF3L, RPSA, RPL18, EIF4A2, RPL13, YBX1, EIF3E, KHDRBS1 |
903 |
| GOBP_FIBROBLAST_PROLIFERATION | 3.87e-03 | 10.36 | 2.02 | 1.00e+00 | 1.00e+00 | 3PEX2, CDK6, MYC |
80 |
| GOBP_NEGATIVE_REGULATION_OF_FIBROBLAST_PROLIFERATION | 6.69e-03 | 17.94 | 2.02 | 1.00e+00 | 1.00e+00 | 2PEX2, MYC |
31 |
Dowload full table
Immunological Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GSE21927_SPLEEN_C57BL6_VS_4T1_TUMOR_BALBC_MONOCYTES_UP | 9.73e-07 | 12.40 | 4.96 | 2.37e-03 | 4.74e-03 | 8EEF1B2, EEF2, NACA, UQCRH, BTF3, EIF3L, RPL18, RPL13 |
200 |
| GSE42088_UNINF_VS_LEISHMANIA_INF_DC_2H_DN | 9.73e-07 | 12.40 | 4.96 | 2.37e-03 | 4.74e-03 | 8EEF2, NACA, POLR1D, BTF3, EIF3L, RPL13, EIF3E, NSA2 |
200 |
| GSE22886_NAIVE_BCELL_VS_NEUTROPHIL_UP | 1.15e-05 | 10.60 | 3.97 | 8.26e-03 | 5.60e-02 | 7NPM1, EEF2, PKIA, HNRNPA1, EIF3L, EIF4A2, TTC3 |
199 |
| GSE24210_CTRL_VS_IL35_TREATED_TCONV_CD4_TCELL_DN | 1.15e-05 | 10.60 | 3.97 | 8.26e-03 | 5.60e-02 | 7NPM1, PEX2, POLR1D, PHB2, YBX1, AHCY, DLL3 |
199 |
| GSE17301_CTRL_VS_48H_ACD3_ACD28_STIM_CD8_TCELL_UP | 1.15e-05 | 10.60 | 3.97 | 8.26e-03 | 5.60e-02 | 7EEF2, HNRNPA1, RPL18, EIF4A2, MYC, PHB2, PPP1R14B |
199 |
| GSE2405_0H_VS_24H_A_PHAGOCYTOPHILUM_STIM_NEUTROPHIL_UP | 1.19e-05 | 10.54 | 3.95 | 8.26e-03 | 5.79e-02 | 7NPM1, EEF1B2, RPSA, RPL18, RPL13, YBX1, EIF3E |
200 |
| GSE25085_FETAL_LIVER_VS_FETAL_BM_SP4_THYMIC_IMPLANT_UP | 1.19e-05 | 10.54 | 3.95 | 8.26e-03 | 5.79e-02 | 7BTF3, ICA1, EIF3L, RPL13, EIF3E, NME2, NSA2 |
200 |
| GSE6269_FLU_VS_E_COLI_INF_PBMC_DN | 4.18e-05 | 10.78 | 3.70 | 2.26e-02 | 2.04e-01 | 6EEF1B2, EEF2, BTF3, EIF3L, CCNI, RPL13 |
164 |
| GSE3565_DUSP1_VS_WT_SPLENOCYTES_POST_LPS_INJECTION_UP | 4.18e-05 | 10.78 | 3.70 | 2.26e-02 | 2.04e-01 | 6NPM1, EEF1B2, RPL18, MYC, RPL13, AHCY |
164 |
| GSE3720_LPS_VS_PMA_STIM_VD1_GAMMADELTA_TCELL_DN | 5.43e-05 | 10.27 | 3.53 | 2.65e-02 | 2.65e-01 | 6EEF1B2, EEF1A1, RPSA, RPL18, MYC, CCNI |
172 |
| GSE3720_UNSTIM_VS_PMA_STIM_VD2_GAMMADELTA_TCELL_UP | 5.98e-05 | 10.09 | 3.46 | 2.65e-02 | 2.91e-01 | 6EEF1B2, NACA, EEF1A1, RPSA, RPL18, PPP1R14B |
175 |
| GSE34205_HEALTHY_VS_FLU_INF_INFANT_PBMC_UP | 1.17e-04 | 8.88 | 3.05 | 3.77e-02 | 5.71e-01 | 6EEF1B2, EEF2, NACA, EIF3L, RPL18, RPL13 |
198 |
| GSE36009_WT_VS_NLRP10_KO_DC_LPS_STIM_DN | 1.20e-04 | 8.83 | 3.04 | 3.77e-02 | 5.86e-01 | 6HNRNPA1, ZC2HC1A, EIF4A2, TTC3, RPL13, NSA2 |
199 |
| GSE42088_UNINF_VS_LEISHMANIA_INF_DC_4H_DN | 1.20e-04 | 8.83 | 3.04 | 3.77e-02 | 5.86e-01 | 6NACA, BTF3, EIF3L, RPL18, RPL13, EIF3E |
199 |
| GSE2405_0H_VS_9H_A_PHAGOCYTOPHILUM_STIM_NEUTROPHIL_DN | 1.24e-04 | 8.78 | 3.02 | 3.77e-02 | 6.03e-01 | 6NPM1, EEF1B2, RPSA, RPL18, RPL13, YBX1 |
200 |
| GSE41978_ID2_KO_VS_ID2_KO_AND_BIM_KO_KLRG1_LOW_EFFECTOR_CD8_TCELL_DN | 1.24e-04 | 8.78 | 3.02 | 3.77e-02 | 6.03e-01 | 6EEF2, RPSA, RPL18, RPL13, YBX1, EIF3E |
200 |
| GSE3565_CTRL_VS_LPS_INJECTED_DUSP1_KO_SPLENOCYTES_DN | 5.38e-04 | 8.37 | 2.56 | 1.49e-01 | 1.00e+00 | 5EEF1B2, EEF2, RPL18, MYC, TTC3 |
171 |
| GSE14415_INDUCED_TREG_VS_FOXP3_KO_INDUCED_TREG_UP | 5.52e-04 | 8.32 | 2.54 | 1.49e-01 | 1.00e+00 | 5EEF1B2, MYC, TTC3, RPL13, EIF3E |
172 |
| GSE3565_CTRL_VS_LPS_INJECTED_SPLENOCYTES_DN | 5.97e-04 | 8.17 | 2.50 | 1.49e-01 | 1.00e+00 | 5EEF1B2, EEF2, RPL18, MYC, RPL13 |
175 |
| GSE3720_UNSTIM_VS_PMA_STIM_VD1_GAMMADELTA_TCELL_UP | 6.12e-04 | 8.12 | 2.49 | 1.49e-01 | 1.00e+00 | 5EEF1B2, NACA, RPSA, MYC, NME2 |
176 |
Top Ranked Transcription Factors for this Gene Expression Program:
| Gene Symbol | TF Rank | DNA Binding Domain | Motif Status | IUPAC PWM | GTEx | DepMap | Decartes |
|---|---|---|---|---|---|---|---|
| NPM1 | 1 | No | Unlikely to be sequence specific TF | Low specificity DNA-binding protein | No motif | None | No evidence for sequence-specific DNA-binding (PMID: 2223875) |
| ZFHX4 | 2 | Yes | Inferred motif | Monomer or homomultimer | In vivo/Misc source | None | Curious protein with numerous C2H2 and homeodomain-like domains. |
| HNRNPA1 | 11 | No | ssDNA/RNA binding | Not a DNA binding protein | No motif | None | Binds single stranded DNA in the structure (PDB: 1PGZ) |
| MEIS2 | 19 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| MYC | 30 | Yes | Known motif | Obligate heteromer | In vivo/Misc source | None | Functions as a heterodimer with MAX. |
| PHB2 | 34 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Protein has been shown to bind to TFs and function as a co-repressor. There appears to be no evidence for direct DNA-binding activity (PMID: 18629613) |
| PHOX2A | 36 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| YBX1 | 40 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | Might also bind RNA |
| NME2 | 42 | Yes | Likely to be sequence specific TF | Monomer or homomultimer | No motif | None | Binds to telomeres (PMID: 22135295). Binds specific sequences by gel-shift, and binding is impacted by a SNP (PMID: 23368879) |
| HMGA1 | 54 | Yes | Known motif | Monomer or homomultimer | In vivo/Misc source | Only known motifs are from Transfac or HocoMoco - origin is uncertain | None |
| SPDEF | 77 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| TRIM28 | 83 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | Only known motifs are from Transfac or HocoMoco - origin is uncertain | This protein a well-known co-repressor and is recruited to DNA by KRAB-domain containing TFs. Transfac motifs are likely from interacting proteins |
| NFATC4 | 84 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| PCBP2 | 91 | No | ssDNA/RNA binding | Not a DNA binding protein | No motif | None | Protein is an RBP. Structure (PDB:2PQU) is with single stranded DNA |
| BMP7 | 116 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| HDAC2 | 122 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | Only known motifs are from Transfac or HocoMoco - origin is uncertain | Histone deacetylase; likely to be a transcriptional cofactor |
| CXXC4 | 123 | Yes | Likely to be sequence specific TF | Monomer or homomultimer | No motif | None | Binds unmethylated CpG-rich sequences: (PMID: 23563267). |
| APEX1 | 125 | No | Unlikely to be sequence specific TF | Low specificity DNA-binding protein | No motif | None | The structure (PDB:1DEW) is with abasic DNA - the protein is a base excision repair enzyme that cleaves off abasic bases |
| ALK | 137 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Operates far upstream in the signaling cascade |
| FOXO3 | 142 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
QQ Plot showing correlations with other GEPs in this dataset, calculated by Spearman correlation:
Interactive QQ-plot of gene loadings:
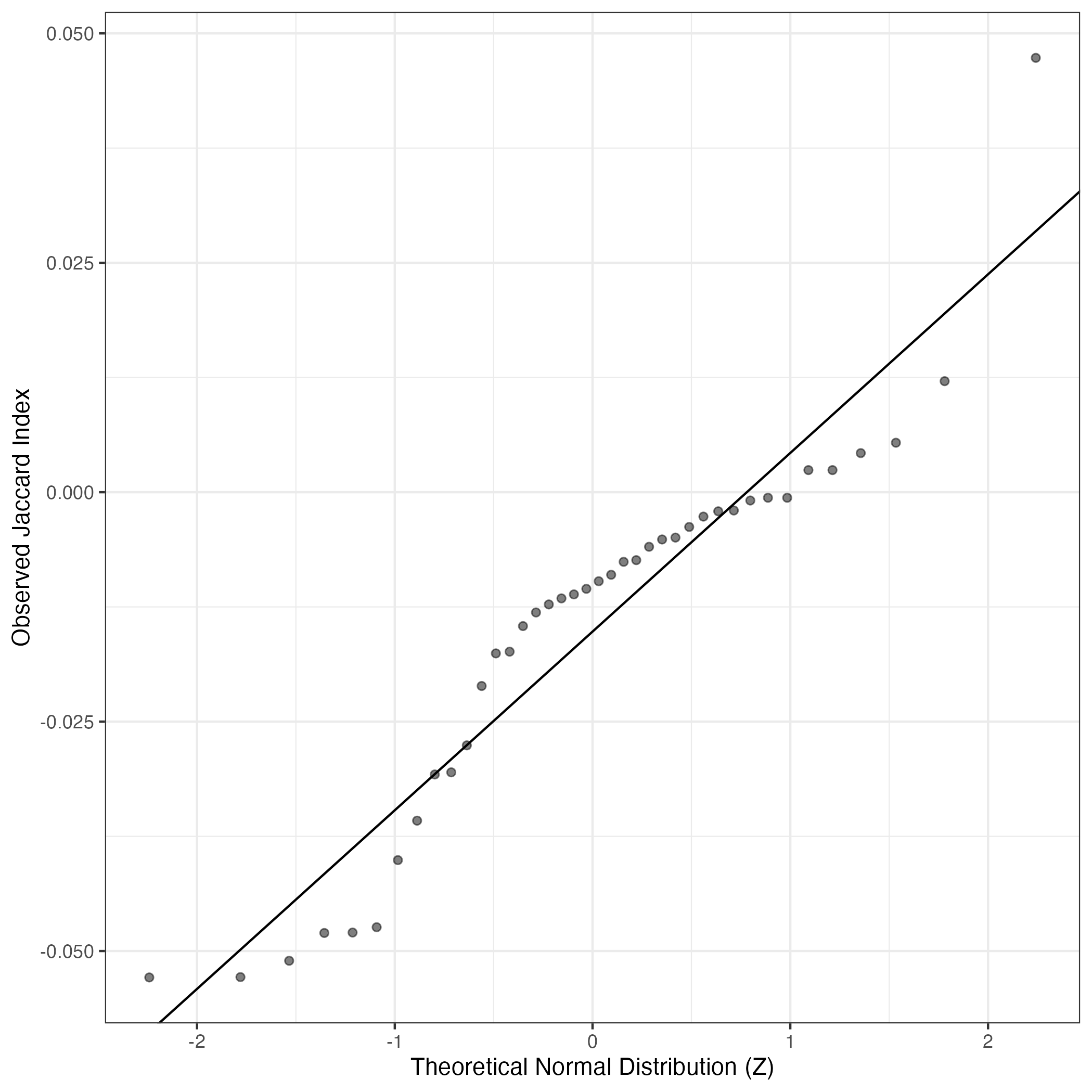
A similar QQ-plot as above, but only for instances where the H value is e.g. > 25, i.e. we are confident that the expression program is active above noise. Agreemenet between these binary vectors is tested using the Jaccard Index, with the P-values calculated by an exact test:
Interactive QQ-plot:
Singler cell type annotations for the top 50 cells on this program.
| Cell ID | Singler label | Singler Delta | Activity Score | Top Singler Raw Scores |
|---|---|---|---|---|
| STDY7787237_AAATGCCTCCACGCAG | Neurons:adrenal_medulla_cell_line | 0.21 | 1278.66 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.53, Neuroepithelial_cell:ESC-derived: 0.44, Astrocyte:Embryonic_stem_cell-derived: 0.43, iPS_cells:PDB_1lox-21Puro-20: 0.41, iPS_cells:PDB_1lox-21Puro-26: 0.41, iPS_cells:PDB_1lox-17Puro-10: 0.41, iPS_cells:PDB_1lox-17Puro-5: 0.41, Neurons:ES_cell-derived_neural_precursor: 0.41, iPS_cells:PDB_2lox-22: 0.4, iPS_cells:PDB_2lox-21: 0.4 |
| STDY7787237_CTGCTGTAGTCGTACT | Neurons:adrenal_medulla_cell_line | 0.25 | 1246.29 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.58, Neuroepithelial_cell:ESC-derived: 0.48, Astrocyte:Embryonic_stem_cell-derived: 0.47, iPS_cells:PDB_1lox-21Puro-20: 0.47, iPS_cells:PDB_1lox-21Puro-26: 0.47, iPS_cells:PDB_1lox-17Puro-10: 0.47, iPS_cells:PDB_1lox-17Puro-5: 0.46, iPS_cells:PDB_2lox-22: 0.46, Embryonic_stem_cells: 0.45, iPS_cells:PDB_2lox-21: 0.45 |
| STDY7787239_AGGTCCGCAGCCTGTG | Neurons:adrenal_medulla_cell_line | 0.23 | 1233.49 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.53, Astrocyte:Embryonic_stem_cell-derived: 0.44, Neuroepithelial_cell:ESC-derived: 0.43, iPS_cells:PDB_1lox-21Puro-20: 0.41, iPS_cells:PDB_1lox-21Puro-26: 0.41, iPS_cells:PDB_1lox-17Puro-10: 0.41, iPS_cells:PDB_1lox-17Puro-5: 0.41, iPS_cells:PDB_2lox-21: 0.4, iPS_cells:PDB_2lox-22: 0.4, Neurons:ES_cell-derived_neural_precursor: 0.4 |
| STDY7787237_TTTGCGCGTACGACCC | Neurons:adrenal_medulla_cell_line | 0.18 | 903.47 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.48, Astrocyte:Embryonic_stem_cell-derived: 0.39, Neuroepithelial_cell:ESC-derived: 0.39, iPS_cells:PDB_1lox-21Puro-20: 0.39, iPS_cells:PDB_1lox-17Puro-10: 0.38, iPS_cells:PDB_1lox-21Puro-26: 0.38, iPS_cells:PDB_1lox-17Puro-5: 0.38, iPS_cells:PDB_2lox-22: 0.38, iPS_cells:PDB_2lox-21: 0.38, iPS_cells:PDB_2lox-17: 0.38 |
| STDY7787238_TACTCGCGTATATGGA | Neurons:adrenal_medulla_cell_line | 0.22 | 755.78 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.51, Neuroepithelial_cell:ESC-derived: 0.43, iPS_cells:PDB_1lox-21Puro-20: 0.43, iPS_cells:PDB_1lox-21Puro-26: 0.42, iPS_cells:PDB_1lox-17Puro-10: 0.42, Astrocyte:Embryonic_stem_cell-derived: 0.42, iPS_cells:PDB_1lox-17Puro-5: 0.42, iPS_cells:PDB_2lox-22: 0.42, iPS_cells:PDB_2lox-21: 0.41, iPS_cells:PDB_2lox-17: 0.41 |
| STDY7787238_GGATTACAGAGGTAGA | Neurons:adrenal_medulla_cell_line | 0.19 | 723.33 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.46, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.37, iPS_cells:PDB_1lox-21Puro-26: 0.36, iPS_cells:PDB_1lox-17Puro-10: 0.36, iPS_cells:PDB_1lox-17Puro-5: 0.36, iPS_cells:PDB_2lox-21: 0.35, iPS_cells:PDB_2lox-22: 0.35, iPS_cells:PDB_2lox-17: 0.35 |
| STDY7787239_TTCTCAATCTCTAAGG | Neurons:adrenal_medulla_cell_line | 0.21 | 685.19 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.47, Neuroepithelial_cell:ESC-derived: 0.37, Astrocyte:Embryonic_stem_cell-derived: 0.37, iPS_cells:PDB_1lox-21Puro-20: 0.36, iPS_cells:PDB_1lox-21Puro-26: 0.36, iPS_cells:PDB_1lox-17Puro-10: 0.36, iPS_cells:PDB_2lox-22: 0.36, iPS_cells:PDB_1lox-17Puro-5: 0.35, iPS_cells:PDB_2lox-21: 0.35, iPS_cells:PDB_2lox-17: 0.35 |
| STDY7787238_CCGGTAGAGAGTAAGG | Neurons:adrenal_medulla_cell_line | 0.19 | 684.19 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.47, Astrocyte:Embryonic_stem_cell-derived: 0.4, Neuroepithelial_cell:ESC-derived: 0.4, iPS_cells:PDB_2lox-22: 0.37, iPS_cells:PDB_1lox-21Puro-20: 0.37, Neurons:ES_cell-derived_neural_precursor: 0.37, iPS_cells:PDB_1lox-17Puro-10: 0.37, iPS_cells:PDB_1lox-21Puro-26: 0.37, iPS_cells:PDB_2lox-17: 0.37, iPS_cells:PDB_2lox-21: 0.37 |
| STDY7787238_AACCATGCAGATGGCA | Neurons:adrenal_medulla_cell_line | 0.20 | 676.94 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.48, Neuroepithelial_cell:ESC-derived: 0.39, iPS_cells:PDB_1lox-21Puro-20: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-21Puro-26: 0.38, iPS_cells:PDB_2lox-22: 0.38, iPS_cells:PDB_1lox-17Puro-5: 0.38, iPS_cells:PDB_1lox-17Puro-10: 0.38, iPS_cells:PDB_2lox-21: 0.38, iPS_cells:PDB_2lox-17: 0.38 |
| STDY7787237_TACGGTACATACGCCG | Neurons:adrenal_medulla_cell_line | 0.18 | 631.22 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.48, Astrocyte:Embryonic_stem_cell-derived: 0.39, Neuroepithelial_cell:ESC-derived: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.38, iPS_cells:PDB_1lox-17Puro-10: 0.37, iPS_cells:PDB_1lox-21Puro-26: 0.37, iPS_cells:PDB_2lox-22: 0.37, iPS_cells:PDB_1lox-17Puro-5: 0.37, iPS_cells:PDB_2lox-21: 0.37, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.37 |
| STDY7787239_GATCGTATCAGGCCCA | Neurons:adrenal_medulla_cell_line | 0.22 | 627.98 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.47, Neuroepithelial_cell:ESC-derived: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.37, iPS_cells:PDB_1lox-21Puro-26: 0.37, iPS_cells:PDB_1lox-17Puro-10: 0.36, iPS_cells:PDB_1lox-17Puro-5: 0.36, iPS_cells:PDB_2lox-22: 0.36, iPS_cells:PDB_2lox-21: 0.36, iPS_cells:PDB_2lox-17: 0.36 |
| STDY7787237_TAAGCGTCATAACCTG | Neurons:adrenal_medulla_cell_line | 0.19 | 587.86 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.47, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.37, iPS_cells:PDB_1lox-21Puro-26: 0.36, iPS_cells:PDB_1lox-17Puro-10: 0.36, iPS_cells:PDB_1lox-17Puro-5: 0.36, iPS_cells:PDB_2lox-22: 0.36, iPS_cells:PDB_2lox-21: 0.36, iPS_cells:PDB_2lox-17: 0.36 |
| STDY7787238_GGAAAGCTCTTCTGGC | Neurons:adrenal_medulla_cell_line | 0.20 | 550.14 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.46, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.36, iPS_cells:PDB_1lox-21Puro-26: 0.36, iPS_cells:PDB_1lox-17Puro-10: 0.36, iPS_cells:PDB_1lox-17Puro-5: 0.35, iPS_cells:PDB_2lox-22: 0.35, iPS_cells:PDB_2lox-21: 0.35, iPS_cells:PDB_2lox-17: 0.35 |
| STDY7787239_ACGTCAACAATCCAAC | Neurons:adrenal_medulla_cell_line | 0.22 | 549.60 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.5, Astrocyte:Embryonic_stem_cell-derived: 0.4, Neuroepithelial_cell:ESC-derived: 0.4, iPS_cells:PDB_1lox-21Puro-20: 0.39, iPS_cells:PDB_1lox-21Puro-26: 0.38, iPS_cells:PDB_1lox-17Puro-10: 0.38, iPS_cells:PDB_1lox-17Puro-5: 0.38, Neurons:ES_cell-derived_neural_precursor: 0.38, iPS_cells:PDB_2lox-22: 0.38, iPS_cells:PDB_2lox-21: 0.37 |
| STDY7787238_CACATTTGTTCTGAAC | Neurons:adrenal_medulla_cell_line | 0.19 | 540.05 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.44, Astrocyte:Embryonic_stem_cell-derived: 0.38, Neuroepithelial_cell:ESC-derived: 0.37, iPS_cells:PDB_1lox-21Puro-20: 0.35, iPS_cells:PDB_1lox-21Puro-26: 0.35, iPS_cells:PDB_2lox-22: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.35, iPS_cells:PDB_2lox-21: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.35, iPS_cells:PDB_2lox-17: 0.35 |
| STDY7787239_CCTTACGTCACTCCTG | Neurons:adrenal_medulla_cell_line | 0.20 | 530.96 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.46, Astrocyte:Embryonic_stem_cell-derived: 0.38, Neuroepithelial_cell:ESC-derived: 0.37, iPS_cells:PDB_1lox-21Puro-20: 0.35, iPS_cells:PDB_1lox-21Puro-26: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.35, Neurons:ES_cell-derived_neural_precursor: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.34, iPS_cells:PDB_2lox-21: 0.34, iPS_cells:PDB_2lox-22: 0.34 |
| STDY7787238_CTACGTCGTCCGAAGA | Neurons:adrenal_medulla_cell_line | 0.18 | 524.03 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.43, Astrocyte:Embryonic_stem_cell-derived: 0.35, Neuroepithelial_cell:ESC-derived: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_2lox-22: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.33, iPS_cells:PDB_2lox-21: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.33, iPS_cells:PDB_2lox-17: 0.33 |
| STDY7787237_AACGTTGTCGATAGAA | Neurons:adrenal_medulla_cell_line | 0.22 | 505.93 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.52, Neuroepithelial_cell:ESC-derived: 0.44, Astrocyte:Embryonic_stem_cell-derived: 0.43, iPS_cells:PDB_1lox-21Puro-20: 0.43, iPS_cells:PDB_1lox-21Puro-26: 0.42, iPS_cells:PDB_2lox-22: 0.42, iPS_cells:PDB_1lox-17Puro-10: 0.42, iPS_cells:PDB_1lox-17Puro-5: 0.42, iPS_cells:PDB_2lox-21: 0.42, iPS_cells:PDB_2lox-17: 0.42 |
| STDY7787239_CGGTTAACAAGGCTCC | Neurons:adrenal_medulla_cell_line | 0.21 | 485.75 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.48, Neuroepithelial_cell:ESC-derived: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.37, iPS_cells:PDB_1lox-21Puro-26: 0.37, iPS_cells:PDB_1lox-17Puro-10: 0.37, Neurons:ES_cell-derived_neural_precursor: 0.37, iPS_cells:PDB_1lox-17Puro-5: 0.37, iPS_cells:PDB_2lox-21: 0.37, iPS_cells:PDB_2lox-22: 0.36 |
| STDY7787238_CTGTGCTAGTGAAGAG | Neurons:adrenal_medulla_cell_line | 0.18 | 481.97 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.43, Neuroepithelial_cell:ESC-derived: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.33, iPS_cells:PDB_2lox-22: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.33, iPS_cells:PDB_2lox-21: 0.33, iPS_cells:PDB_2lox-17: 0.33 |
| STDY7787237_TCGCGAGTCAGCTGGC | Neurons:adrenal_medulla_cell_line | 0.16 | 470.93 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.41, Astrocyte:Embryonic_stem_cell-derived: 0.34, Neuroepithelial_cell:ESC-derived: 0.34, iPS_cells:PDB_1lox-21Puro-20: 0.32, iPS_cells:PDB_1lox-21Puro-26: 0.32, iPS_cells:PDB_1lox-17Puro-10: 0.32, iPS_cells:PDB_1lox-17Puro-5: 0.32, iPS_cells:PDB_2lox-21: 0.32, iPS_cells:PDB_2lox-22: 0.32, iPS_cells:PDB_2lox-17: 0.32 |
| STDY7787239_GGAACTTAGGGCACTA | Neurons:adrenal_medulla_cell_line | 0.20 | 433.72 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.44, Neuroepithelial_cell:ESC-derived: 0.37, Astrocyte:Embryonic_stem_cell-derived: 0.37, iPS_cells:PDB_1lox-21Puro-20: 0.35, Neurons:ES_cell-derived_neural_precursor: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.34, iPS_cells:PDB_2lox-22: 0.34, iPS_cells:PDB_2lox-21: 0.34 |
| STDY7787239_GACGTTACATGGTAGG | Neurons:adrenal_medulla_cell_line | 0.20 | 431.95 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.43, Neuroepithelial_cell:ESC-derived: 0.36, Astrocyte:Embryonic_stem_cell-derived: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.35, iPS_cells:PDB_1lox-21Puro-26: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.34, iPS_cells:PDB_2lox-22: 0.34, iPS_cells:PDB_2lox-21: 0.34, iPS_cells:PDB_2lox-17: 0.34 |
| STDY7787238_GGTGTTACAATAGAGT | Neurons:adrenal_medulla_cell_line | 0.18 | 430.21 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.41, Neuroepithelial_cell:ESC-derived: 0.34, iPS_cells:PDB_1lox-21Puro-20: 0.33, iPS_cells:PDB_1lox-21Puro-26: 0.33, Astrocyte:Embryonic_stem_cell-derived: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.33, iPS_cells:PDB_2lox-22: 0.33, iPS_cells:PDB_2lox-21: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.33, iPS_cells:PDB_2lox-17: 0.33 |
| STDY7787238_GGATTACGTGCGCTTG | Neurons:adrenal_medulla_cell_line | 0.15 | 400.02 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.32, Neuroepithelial_cell:ESC-derived: 0.3, iPS_cells:PDB_1lox-21Puro-20: 0.3, iPS_cells:PDB_1lox-21Puro-26: 0.3, iPS_cells:PDB_1lox-17Puro-10: 0.29, iPS_cells:PDB_1lox-17Puro-5: 0.29, iPS_cells:PDB_2lox-22: 0.29, iPS_cells:PDB_2lox-21: 0.29, iPS_cells:iPS:minicircle-derived: 0.29 |
| STDY7787237_CACATAGTCTATGTGG | Neurons:adrenal_medulla_cell_line | 0.17 | 398.94 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.43, Neuroepithelial_cell:ESC-derived: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_2lox-22: 0.34, iPS_cells:PDB_2lox-21: 0.34, iPS_cells:PDB_2lox-17: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.34 |
| STDY7787238_GTTACAGAGCAAATCA | Neurons:adrenal_medulla_cell_line | 0.18 | 388.70 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.43, Neuroepithelial_cell:ESC-derived: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.33, iPS_cells:PDB_2lox-22: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.33, iPS_cells:PDB_2lox-21: 0.33, iPS_cells:PDB_2lox-17: 0.33 |
| STDY7787239_ATGTGTGAGTCCTCCT | Neurons:adrenal_medulla_cell_line | 0.18 | 375.92 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.41, Neuroepithelial_cell:ESC-derived: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.34, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.33, iPS_cells:PDB_1lox-21Puro-26: 0.33, iPS_cells:PDB_2lox-22: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.33, iPS_cells:PDB_2lox-17: 0.33, iPS_cells:PDB_2lox-21: 0.33 |
| STDY7787237_TTCTACAAGCGTCTAT | Neurons:adrenal_medulla_cell_line | 0.19 | 370.93 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.47, Neuroepithelial_cell:ESC-derived: 0.41, Astrocyte:Embryonic_stem_cell-derived: 0.41, iPS_cells:PDB_1lox-21Puro-20: 0.39, iPS_cells:PDB_1lox-21Puro-26: 0.39, iPS_cells:PDB_1lox-17Puro-10: 0.39, iPS_cells:PDB_1lox-17Puro-5: 0.39, Neurons:ES_cell-derived_neural_precursor: 0.38, iPS_cells:PDB_2lox-22: 0.38, iPS_cells:PDB_2lox-21: 0.38 |
| STDY7787239_GGTATTGGTTGTACAC | Neurons:adrenal_medulla_cell_line | 0.19 | 369.41 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.43, Neuroepithelial_cell:ESC-derived: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.34, iPS_cells:PDB_2lox-22: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_2lox-21: 0.33, iPS_cells:PDB_2lox-17: 0.33 |
| STDY7787237_ACGGGTCAGAAGGTGA | Neurons:adrenal_medulla_cell_line | 0.19 | 360.48 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.44, Astrocyte:Embryonic_stem_cell-derived: 0.37, Neuroepithelial_cell:ESC-derived: 0.37, iPS_cells:PDB_1lox-21Puro-20: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_2lox-22: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.34, Neurons:ES_cell-derived_neural_precursor: 0.34, iPS_cells:PDB_2lox-5: 0.34 |
| STDY7787239_TCAGCAAAGGCTAGCA | Neurons:adrenal_medulla_cell_line | 0.17 | 357.13 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.39, Neuroepithelial_cell:ESC-derived: 0.32, Astrocyte:Embryonic_stem_cell-derived: 0.32, iPS_cells:PDB_1lox-21Puro-26: 0.31, iPS_cells:PDB_1lox-21Puro-20: 0.31, iPS_cells:PDB_2lox-21: 0.31, iPS_cells:PDB_1lox-17Puro-5: 0.31, iPS_cells:PDB_2lox-22: 0.31, iPS_cells:PDB_1lox-17Puro-10: 0.31, iPS_cells:PDB_2lox-17: 0.31 |
| STDY7787238_CTAGAGTTCTGAGTGT | Neurons:adrenal_medulla_cell_line | 0.23 | 352.58 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.5, Neuroepithelial_cell:ESC-derived: 0.44, Astrocyte:Embryonic_stem_cell-derived: 0.42, iPS_cells:PDB_1lox-21Puro-20: 0.41, iPS_cells:PDB_1lox-21Puro-26: 0.41, iPS_cells:PDB_1lox-17Puro-10: 0.41, iPS_cells:PDB_1lox-17Puro-5: 0.41, iPS_cells:PDB_2lox-22: 0.41, Neurons:ES_cell-derived_neural_precursor: 0.4, iPS_cells:PDB_2lox-21: 0.4 |
| STDY7787238_GACTGCGCAATCCAAC | Neurons:adrenal_medulla_cell_line | 0.17 | 342.89 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.41, Astrocyte:Embryonic_stem_cell-derived: 0.34, Neuroepithelial_cell:ESC-derived: 0.33, iPS_cells:PDB_1lox-21Puro-20: 0.32, iPS_cells:PDB_1lox-21Puro-26: 0.32, iPS_cells:PDB_1lox-17Puro-5: 0.31, iPS_cells:PDB_1lox-17Puro-10: 0.31, iPS_cells:PDB_2lox-22: 0.31, iPS_cells:PDB_2lox-21: 0.31, iPS_cells:PDB_2lox-17: 0.31 |
| STDY7787239_CTTACCGTCACTTATC | Neurons:adrenal_medulla_cell_line | 0.24 | 340.35 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.5, Neuroepithelial_cell:ESC-derived: 0.42, Astrocyte:Embryonic_stem_cell-derived: 0.41, iPS_cells:PDB_1lox-21Puro-20: 0.4, iPS_cells:PDB_1lox-21Puro-26: 0.39, iPS_cells:PDB_1lox-17Puro-10: 0.39, iPS_cells:PDB_1lox-17Puro-5: 0.39, Neurons:ES_cell-derived_neural_precursor: 0.39, iPS_cells:PDB_2lox-22: 0.39, Embryonic_stem_cells: 0.39 |
| STDY7787237_TACGGGCGTTCACCTC | Neurons:adrenal_medulla_cell_line | 0.18 | 321.03 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.44, Astrocyte:Embryonic_stem_cell-derived: 0.35, Neuroepithelial_cell:ESC-derived: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_2lox-22: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.34, iPS_cells:PDB_2lox-21: 0.34, iPS_cells:PDB_2lox-17: 0.34 |
| STDY7787239_TCTCTAACAACACCCG | Neurons:adrenal_medulla_cell_line | 0.25 | 320.28 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.5, Neuroepithelial_cell:ESC-derived: 0.43, Astrocyte:Embryonic_stem_cell-derived: 0.41, iPS_cells:PDB_1lox-21Puro-20: 0.41, iPS_cells:PDB_1lox-21Puro-26: 0.41, iPS_cells:PDB_1lox-17Puro-10: 0.41, iPS_cells:PDB_1lox-17Puro-5: 0.4, iPS_cells:PDB_2lox-22: 0.4, Neurons:ES_cell-derived_neural_precursor: 0.4, iPS_cells:PDB_2lox-21: 0.4 |
| STDY7787237_ATCCGAAAGGTGACCA | Neurons:adrenal_medulla_cell_line | 0.17 | 318.28 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.41, Astrocyte:Embryonic_stem_cell-derived: 0.34, Neuroepithelial_cell:ESC-derived: 0.33, iPS_cells:PDB_1lox-21Puro-20: 0.32, iPS_cells:PDB_1lox-17Puro-10: 0.32, iPS_cells:PDB_1lox-21Puro-26: 0.32, iPS_cells:PDB_2lox-22: 0.32, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.32, iPS_cells:PDB_1lox-17Puro-5: 0.32, iPS_cells:PDB_2lox-21: 0.31 |
| STDY7787238_CGAGCACTCCAAAGTC | Neurons:adrenal_medulla_cell_line | 0.20 | 314.66 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.45, Neuroepithelial_cell:ESC-derived: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.37, iPS_cells:PDB_1lox-21Puro-26: 0.37, iPS_cells:PDB_2lox-22: 0.37, iPS_cells:PDB_1lox-17Puro-5: 0.37, iPS_cells:PDB_1lox-17Puro-10: 0.36, Astrocyte:Embryonic_stem_cell-derived: 0.36, iPS_cells:PDB_2lox-21: 0.36, iPS_cells:PDB_2lox-17: 0.36 |
| STDY7787239_CCAGCGACATCCTTGC | Neurons:adrenal_medulla_cell_line | 0.25 | 314.29 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.48, Neuroepithelial_cell:ESC-derived: 0.39, iPS_cells:PDB_1lox-21Puro-20: 0.38, iPS_cells:PDB_1lox-21Puro-26: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-17Puro-10: 0.37, iPS_cells:PDB_1lox-17Puro-5: 0.37, Neurons:ES_cell-derived_neural_precursor: 0.37, Embryonic_stem_cells: 0.36, iPS_cells:PDB_2lox-22: 0.36 |
| STDY7787238_AAGGTTCCATAGTAAG | Neurons:adrenal_medulla_cell_line | 0.18 | 313.47 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.32, Neuroepithelial_cell:ESC-derived: 0.32, iPS_cells:PDB_1lox-21Puro-20: 0.31, iPS_cells:PDB_1lox-17Puro-10: 0.3, iPS_cells:PDB_1lox-21Puro-26: 0.3, iPS_cells:PDB_1lox-17Puro-5: 0.3, iPS_cells:PDB_2lox-22: 0.3, iPS_cells:PDB_2lox-21: 0.3, iPS_cells:PDB_2lox-17: 0.3 |
| STDY7787238_CAAGATCGTCCGAACC | Neurons:adrenal_medulla_cell_line | 0.22 | 309.58 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.49, Astrocyte:Embryonic_stem_cell-derived: 0.43, Neuroepithelial_cell:ESC-derived: 0.42, Neurons:ES_cell-derived_neural_precursor: 0.39, iPS_cells:PDB_1lox-21Puro-20: 0.39, iPS_cells:PDB_1lox-21Puro-26: 0.39, iPS_cells:PDB_1lox-17Puro-10: 0.38, iPS_cells:PDB_2lox-22: 0.38, iPS_cells:PDB_2lox-21: 0.38, iPS_cells:PDB_1lox-17Puro-5: 0.38 |
| STDY7787238_TCGCGAGAGATGAGAG | Neurons:adrenal_medulla_cell_line | 0.17 | 307.54 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.3, Neuroepithelial_cell:ESC-derived: 0.3, iPS_cells:PDB_1lox-21Puro-20: 0.29, iPS_cells:PDB_1lox-17Puro-5: 0.29, iPS_cells:PDB_1lox-21Puro-26: 0.29, iPS_cells:PDB_1lox-17Puro-10: 0.29, iPS_cells:PDB_2lox-22: 0.29, iPS_cells:PDB_2lox-21: 0.28, iPS_cells:PDB_2lox-17: 0.28 |
| STDY7787237_CGGTTAACAGGAACGT | Neurons:adrenal_medulla_cell_line | 0.14 | 302.61 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.32, Neuroepithelial_cell:ESC-derived: 0.32, iPS_cells:PDB_1lox-21Puro-20: 0.31, iPS_cells:PDB_1lox-21Puro-26: 0.31, iPS_cells:PDB_1lox-17Puro-5: 0.31, iPS_cells:PDB_1lox-17Puro-10: 0.31, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.31, iPS_cells:PDB_2lox-22: 0.3, iPS_cells:PDB_2lox-21: 0.3 |
| STDY7787237_TTGCGTCCAATGGAGC | Neurons:adrenal_medulla_cell_line | 0.16 | 299.97 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.41, Astrocyte:Embryonic_stem_cell-derived: 0.35, Neuroepithelial_cell:ESC-derived: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.33, iPS_cells:PDB_1lox-21Puro-26: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.33, Neurons:ES_cell-derived_neural_precursor: 0.32, iPS_cells:PDB_2lox-22: 0.32, iPS_cells:PDB_2lox-21: 0.32 |
| STDY7787237_GTACGTATCCTAGGGC | Neurons:adrenal_medulla_cell_line | 0.18 | 298.46 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.43, Neuroepithelial_cell:ESC-derived: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.34, iPS_cells:PDB_2lox-22: 0.33, iPS_cells:PDB_2lox-21: 0.33, iPS_cells:iPS:minicircle-derived: 0.33 |
| STDY7787237_GGGCACTGTATAGTAG | Neurons:adrenal_medulla_cell_line | 0.14 | 295.76 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.36, Neuroepithelial_cell:ESC-derived: 0.29, Astrocyte:Embryonic_stem_cell-derived: 0.29, iPS_cells:PDB_1lox-21Puro-20: 0.28, iPS_cells:PDB_1lox-21Puro-26: 0.28, iPS_cells:adipose_stem_cell-derived:lentiviral: 0.27, iPS_cells:PDB_1lox-17Puro-5: 0.27, iPS_cells:PDB_1lox-17Puro-10: 0.27, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.27, iPS_cells:PDB_2lox-17: 0.27 |
| STDY7787238_CTGCGGACATGAGCGA | Neurons:adrenal_medulla_cell_line | 0.18 | 294.03 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.41, Neuroepithelial_cell:ESC-derived: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.34, Neurons:ES_cell-derived_neural_precursor: 0.33, iPS_cells:PDB_1lox-21Puro-20: 0.33, iPS_cells:PDB_1lox-21Puro-26: 0.32, iPS_cells:PDB_2lox-22: 0.32, iPS_cells:PDB_1lox-17Puro-10: 0.32, iPS_cells:PDB_2lox-17: 0.32, iPS_cells:PDB_2lox-21: 0.32 |
| STDY7787238_GTTTCTAGTGAAAGAG | Neurons:adrenal_medulla_cell_line | 0.22 | 291.05 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.47, Neuroepithelial_cell:ESC-derived: 0.4, Astrocyte:Embryonic_stem_cell-derived: 0.4, iPS_cells:PDB_1lox-21Puro-20: 0.39, iPS_cells:PDB_1lox-21Puro-26: 0.38, iPS_cells:PDB_1lox-17Puro-10: 0.38, iPS_cells:PDB_1lox-17Puro-5: 0.38, iPS_cells:PDB_2lox-22: 0.38, Embryonic_stem_cells: 0.38, iPS_cells:PDB_2lox-21: 0.38 |
| STDY7787239_GTCATTTCATACTACG | Neurons:adrenal_medulla_cell_line | 0.19 | 290.59 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.44, Neuroepithelial_cell:ESC-derived: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_2lox-21: 0.34, iPS_cells:PDB_2lox-22: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.33, iPS_cells:iPS:minicircle-derived: 0.33 |
Below shows the significant enrichments of this GEP for literature curated gene lists
This data was procured from existing single cell RNA-seq maps of neuroblastoma or related relevant data.
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by the P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Symphathoblasts (Kameneva)
Marker gene were obtained from Fig. 1D of Kameneva et al (PMID 33833454). These genes were used by the authors to annotate each cell type in their human fetal adrenal scRNA-seq data obtained 6, 8, 9, 11, 12 and 14 weeks post conception. Several of these markers are also (more weakly) expressed in chromaffin cells. The authors data suggest Symphthoblasts give rise to chromaffin cells.:
Wilcoxon ranksum test P-value for gene set overrepresentation: 6.11e-04
Mean rank of genes in gene set: 418.5
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ELAVL3 | 0.0032317 | 127 | GTEx | DepMap | Descartes | 3.44 | 154.55 |
| ISL1 | 0.0024248 | 253 | GTEx | DepMap | Descartes | 3.02 | 317.97 |
| STMN2 | 0.0022958 | 275 | GTEx | DepMap | Descartes | 60.80 | 6980.94 |
| ELAVL4 | 0.0009324 | 1019 | GTEx | DepMap | Descartes | 6.58 | 358.53 |
Symphathoblasts Fig2 (Kamenva)
Marker genes were obtained from Fig. 2H of Kameneva et al (PMID 33833454) for symphathoblasts (differentiating from SCPs):
Wilcoxon ranksum test P-value for gene set overrepresentation: 7.12e-04
Mean rank of genes in gene set: 1107.8
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ISL1 | 0.0024248 | 253 | GTEx | DepMap | Descartes | 3.02 | 317.97 |
| STMN2 | 0.0022958 | 275 | GTEx | DepMap | Descartes | 60.80 | 6980.94 |
| HAND2 | 0.0022286 | 292 | GTEx | DepMap | Descartes | 5.40 | 485.67 |
| ELAVL4 | 0.0009324 | 1019 | GTEx | DepMap | Descartes | 6.58 | 358.53 |
| ELAVL2 | 0.0001330 | 3700 | GTEx | DepMap | Descartes | 1.70 | 86.72 |
N Chromafin (Hanemaaijer)
Marker genes obtained from Supplementary Table SD of Hanemaaijer et al (PMID 33500353). The authors generated single-cell RNA-seq data (sort-seq, 2,229 cells total) from mouse adrenal glads at E13.5, E14.5, E17.5, E18.5, P1 and P5. These were marker genes that matched with a similar dataset generated by Furlan et al (PMID 28684471). This particular set of markers are for the Norepinepherine Chromaffin subcluster, which is part of the Adrenal Medulla cluster.:
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.04e-03
Mean rank of genes in gene set: 3487.69
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| SCG5 | 0.0027304 | 185 | GTEx | DepMap | Descartes | 3.58 | 774.29 |
| SCG3 | 0.0017089 | 467 | GTEx | DepMap | Descartes | 2.72 | 218.33 |
| SLCO3A1 | 0.0013775 | 641 | GTEx | DepMap | Descartes | 2.04 | 101.04 |
| CACNA2D1 | 0.0012416 | 725 | GTEx | DepMap | Descartes | 0.95 | 38.04 |
| ADCYAP1R1 | 0.0011640 | 793 | GTEx | DepMap | Descartes | 0.24 | 8.56 |
| GNAS | 0.0007491 | 1300 | GTEx | DepMap | Descartes | 18.03 | 1001.51 |
| PPFIA2 | 0.0007486 | 1302 | GTEx | DepMap | Descartes | 0.29 | 14.76 |
| CELF4 | 0.0006602 | 1493 | GTEx | DepMap | Descartes | 3.17 | 185.50 |
| NAP1L5 | 0.0004350 | 2147 | GTEx | DepMap | Descartes | 0.79 | 110.95 |
| SNAP25 | 0.0004229 | 2189 | GTEx | DepMap | Descartes | 2.70 | 250.97 |
| LGR5 | -0.0000332 | 5298 | GTEx | DepMap | Descartes | 0.01 | 0.38 |
| PCLO | -0.0000656 | 5806 | GTEx | DepMap | Descartes | 0.86 | 9.56 |
| SYN2 | -0.0001152 | 6610 | GTEx | DepMap | Descartes | 0.04 | 1.97 |
| CXCL14 | -0.0001422 | 7051 | GTEx | DepMap | Descartes | 0.12 | 15.17 |
| PTPRN | -0.0002042 | 8000 | GTEx | DepMap | Descartes | 0.43 | 14.22 |
| C1QL1 | -0.0007154 | 11796 | GTEx | DepMap | Descartes | 0.13 | 11.69 |
Below shows ranks on this GEP for literature curated gene lists for large gene sets
These include those reported as mesenchymal/adrenergic by Van Groningen et al.
High ranks indicate this gene is a driver of this GEP (note these results are not ordered).
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
VanGroningen Adrenergic Genes
Adrenergic marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.02e-21
Mean rank of genes in gene set: 4327.93
Median rank of genes in gene set: 2297
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| PKIA | 0.0071768 | 8 | GTEx | DepMap | Descartes | 8.91 | 506.47 |
| ICA1 | 0.0064615 | 16 | GTEx | DepMap | Descartes | 7.34 | 795.28 |
| GAL | 0.0054785 | 26 | GTEx | DepMap | Descartes | 17.53 | 5514.95 |
| GLCCI1 | 0.0050009 | 32 | GTEx | DepMap | Descartes | 5.04 | 261.82 |
| CCNI | 0.0049622 | 33 | GTEx | DepMap | Descartes | 22.13 | 1804.42 |
| PHOX2A | 0.0048503 | 36 | GTEx | DepMap | Descartes | 7.94 | 1160.99 |
| OLA1 | 0.0044593 | 51 | GTEx | DepMap | Descartes | 5.42 | 284.70 |
| BMPR1B | 0.0044232 | 53 | GTEx | DepMap | Descartes | 3.29 | 169.35 |
| HMGA1 | 0.0043946 | 54 | GTEx | DepMap | Descartes | 6.60 | 687.19 |
| DDC | 0.0042436 | 61 | GTEx | DepMap | Descartes | 4.00 | 462.11 |
| SYT1 | 0.0041890 | 65 | GTEx | DepMap | Descartes | 6.61 | 331.29 |
| RBP1 | 0.0041687 | 68 | GTEx | DepMap | Descartes | 8.31 | 1011.19 |
| GGCT | 0.0040951 | 70 | GTEx | DepMap | Descartes | 4.76 | 947.37 |
| ARHGEF7 | 0.0040467 | 72 | GTEx | DepMap | Descartes | 4.81 | 176.34 |
| IRS2 | 0.0039453 | 76 | GTEx | DepMap | Descartes | 7.86 | 246.66 |
| REC8 | 0.0037610 | 85 | GTEx | DepMap | Descartes | 2.09 | 177.96 |
| NELL2 | 0.0036839 | 95 | GTEx | DepMap | Descartes | 2.87 | 182.66 |
| GABRB3 | 0.0034401 | 111 | GTEx | DepMap | Descartes | 1.28 | 50.58 |
| BMP7 | 0.0033398 | 116 | GTEx | DepMap | Descartes | 0.67 | 39.46 |
| KLHL23 | 0.0032602 | 124 | GTEx | DepMap | Descartes | 3.09 | 173.36 |
| ELAVL3 | 0.0032317 | 127 | GTEx | DepMap | Descartes | 3.44 | 154.55 |
| ATCAY | 0.0031853 | 134 | GTEx | DepMap | Descartes | 1.83 | 81.30 |
| TMEM97 | 0.0031685 | 136 | GTEx | DepMap | Descartes | 1.80 | 153.62 |
| ALK | 0.0031661 | 137 | GTEx | DepMap | Descartes | 0.52 | 22.11 |
| FOXO3 | 0.0031167 | 142 | GTEx | DepMap | Descartes | 3.70 | 134.55 |
| TFAP2B | 0.0030848 | 144 | GTEx | DepMap | Descartes | 4.31 | 209.60 |
| POPDC3 | 0.0029123 | 163 | GTEx | DepMap | Descartes | 0.37 | 45.68 |
| GRIA2 | 0.0028485 | 171 | GTEx | DepMap | Descartes | 2.07 | 106.18 |
| LRRTM2 | 0.0028439 | 172 | GTEx | DepMap | Descartes | 0.90 | 37.97 |
| FKBP4 | 0.0027984 | 176 | GTEx | DepMap | Descartes | 2.68 | 183.30 |
VanGroningen Mesenchymal Genes
Mesenchymal marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 9024.03
Median rank of genes in gene set: 9941
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ENAH | 0.0033982 | 112 | GTEx | DepMap | Descartes | 3.09 | 55.84 |
| MEST | 0.0030756 | 146 | GTEx | DepMap | Descartes | 2.61 | 265.21 |
| GAS2 | 0.0025010 | 233 | GTEx | DepMap | Descartes | 0.09 | 7.11 |
| LAMB1 | 0.0018501 | 420 | GTEx | DepMap | Descartes | 1.02 | 48.39 |
| NES | 0.0017679 | 443 | GTEx | DepMap | Descartes | 1.51 | 70.46 |
| COL11A1 | 0.0017303 | 455 | GTEx | DepMap | Descartes | 0.39 | 12.28 |
| CTNNA1 | 0.0017150 | 462 | GTEx | DepMap | Descartes | 1.68 | 104.23 |
| ALDH1A3 | 0.0016642 | 494 | GTEx | DepMap | Descartes | 0.28 | 22.67 |
| TMEFF2 | 0.0011092 | 834 | GTEx | DepMap | Descartes | 1.76 | 134.46 |
| PDE3A | 0.0010108 | 935 | GTEx | DepMap | Descartes | 0.13 | 4.05 |
| SCRG1 | 0.0009885 | 964 | GTEx | DepMap | Descartes | 0.55 | 44.85 |
| STAT1 | 0.0008947 | 1073 | GTEx | DepMap | Descartes | 2.44 | 147.94 |
| CTDSP2 | 0.0008810 | 1095 | GTEx | DepMap | Descartes | 0.96 | 49.83 |
| SERPINE2 | 0.0008447 | 1146 | GTEx | DepMap | Descartes | 1.58 | 64.65 |
| FZD1 | 0.0008168 | 1188 | GTEx | DepMap | Descartes | 0.19 | 6.16 |
| CALD1 | 0.0008104 | 1200 | GTEx | DepMap | Descartes | 3.33 | 189.33 |
| KDM5B | 0.0007852 | 1239 | GTEx | DepMap | Descartes | 0.98 | 25.63 |
| TPM1 | 0.0007640 | 1272 | GTEx | DepMap | Descartes | 2.28 | 137.85 |
| CKAP4 | 0.0005651 | 1733 | GTEx | DepMap | Descartes | 1.18 | 94.91 |
| TJP1 | 0.0005365 | 1814 | GTEx | DepMap | Descartes | 0.45 | 13.09 |
| JAM3 | 0.0005289 | 1836 | GTEx | DepMap | Descartes | 0.26 | 16.93 |
| SLC39A14 | 0.0005038 | 1913 | GTEx | DepMap | Descartes | 0.24 | 11.83 |
| SSBP4 | 0.0004793 | 1994 | GTEx | DepMap | Descartes | 1.58 | 210.50 |
| NFIC | 0.0004681 | 2033 | GTEx | DepMap | Descartes | 0.65 | 18.31 |
| LMAN1 | 0.0004525 | 2093 | GTEx | DepMap | Descartes | 1.44 | 75.88 |
| ATP1B1 | 0.0003923 | 2313 | GTEx | DepMap | Descartes | 4.59 | 336.13 |
| PRDX6 | 0.0003497 | 2491 | GTEx | DepMap | Descartes | 2.61 | 335.91 |
| HIBADH | 0.0003406 | 2523 | GTEx | DepMap | Descartes | 0.46 | 64.37 |
| TPM2 | 0.0002914 | 2728 | GTEx | DepMap | Descartes | 0.90 | 145.03 |
| FKBP14 | 0.0002764 | 2807 | GTEx | DepMap | Descartes | 0.13 | 2.96 |
Descartes adrenocortical markers
Top 50 marker genes of adrenocortical cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/adrenocortical/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.59e-01
Mean rank of genes in gene set: 7323.42
Median rank of genes in gene set: 7738.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| GSTA4 | 0.0009560 | 998 | GTEx | DepMap | Descartes | 1.38 | 188.60 |
| SLC1A2 | 0.0008397 | 1155 | GTEx | DepMap | Descartes | 0.27 | 6.16 |
| JAKMIP2 | 0.0006141 | 1612 | GTEx | DepMap | Descartes | 0.65 | 14.12 |
| SLC16A9 | 0.0004838 | 1974 | GTEx | DepMap | Descartes | 0.19 | 10.55 |
| FRMD5 | 0.0004785 | 1997 | GTEx | DepMap | Descartes | 0.10 | 4.93 |
| GRAMD1B | 0.0002553 | 2931 | GTEx | DepMap | Descartes | 0.11 | 3.65 |
| SCARB1 | 0.0002271 | 3079 | GTEx | DepMap | Descartes | 0.41 | 17.65 |
| PDE10A | 0.0002000 | 3253 | GTEx | DepMap | Descartes | 0.10 | 3.20 |
| SGCZ | -0.0000086 | 5037 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SCAP | -0.0000236 | 5176 | GTEx | DepMap | Descartes | 0.20 | 12.32 |
| FREM2 | -0.0000422 | 5424 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TM7SF2 | -0.0000476 | 5501 | GTEx | DepMap | Descartes | 0.34 | 35.16 |
| BAIAP2L1 | -0.0000559 | 5629 | GTEx | DepMap | Descartes | 0.02 | 2.20 |
| MSMO1 | -0.0000651 | 5795 | GTEx | DepMap | Descartes | 0.43 | 39.45 |
| DHCR24 | -0.0000677 | 5833 | GTEx | DepMap | Descartes | 0.38 | 12.77 |
| DHCR7 | -0.0001155 | 6617 | GTEx | DepMap | Descartes | 0.19 | 16.35 |
| HMGCR | -0.0001229 | 6742 | GTEx | DepMap | Descartes | 0.27 | 11.24 |
| IGF1R | -0.0001416 | 7037 | GTEx | DepMap | Descartes | 0.30 | 5.74 |
| CLU | -0.0002339 | 8440 | GTEx | DepMap | Descartes | 3.82 | 364.57 |
| SH3PXD2B | -0.0002633 | 8828 | GTEx | DepMap | Descartes | 0.05 | 1.42 |
| NPC1 | -0.0002766 | 8990 | GTEx | DepMap | Descartes | 0.10 | 3.96 |
| STAR | -0.0002770 | 8994 | GTEx | DepMap | Descartes | 0.07 | 5.42 |
| PEG3 | -0.0003578 | 9922 | GTEx | DepMap | Descartes | 0.21 | NA |
| PAPSS2 | -0.0003694 | 10012 | GTEx | DepMap | Descartes | 0.01 | 0.06 |
| LDLR | -0.0003932 | 10203 | GTEx | DepMap | Descartes | 0.10 | 4.62 |
| INHA | -0.0003972 | 10236 | GTEx | DepMap | Descartes | 0.01 | 0.96 |
| APOC1 | -0.0004073 | 10313 | GTEx | DepMap | Descartes | 2.63 | 282.81 |
| ERN1 | -0.0004268 | 10485 | GTEx | DepMap | Descartes | 0.06 | 1.46 |
| POR | -0.0004983 | 10909 | GTEx | DepMap | Descartes | 0.28 | 25.62 |
| CYB5B | -0.0005300 | 11079 | GTEx | DepMap | Descartes | 0.50 | 29.27 |
Descartes chromaffin markers
Top 50 marker genes of chromaffin cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/chromaffin/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.07e-03
Mean rank of genes in gene set: 4540.76
Median rank of genes in gene set: 2209
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| IL7 | 0.0070711 | 9 | GTEx | DepMap | Descartes | 8.03 | 947.57 |
| GAL | 0.0054785 | 26 | GTEx | DepMap | Descartes | 17.53 | 5514.95 |
| EYA4 | 0.0036362 | 97 | GTEx | DepMap | Descartes | 1.17 | 56.27 |
| ALK | 0.0031661 | 137 | GTEx | DepMap | Descartes | 0.52 | 22.11 |
| MAB21L2 | 0.0025879 | 218 | GTEx | DepMap | Descartes | 0.88 | 84.32 |
| ISL1 | 0.0024248 | 253 | GTEx | DepMap | Descartes | 3.02 | 317.97 |
| STMN2 | 0.0022958 | 275 | GTEx | DepMap | Descartes | 60.80 | 6980.94 |
| RBFOX1 | 0.0020362 | 344 | GTEx | DepMap | Descartes | 1.18 | 49.91 |
| CCND1 | 0.0016987 | 472 | GTEx | DepMap | Descartes | 8.21 | 523.27 |
| EYA1 | 0.0016763 | 488 | GTEx | DepMap | Descartes | 0.49 | 30.65 |
| SLC44A5 | 0.0015246 | 566 | GTEx | DepMap | Descartes | 0.52 | 28.61 |
| RYR2 | 0.0013797 | 640 | GTEx | DepMap | Descartes | 0.40 | 6.27 |
| SLC6A2 | 0.0013236 | 671 | GTEx | DepMap | Descartes | 0.48 | 31.01 |
| TUBB2B | 0.0011765 | 778 | GTEx | DepMap | Descartes | 19.72 | 2404.78 |
| TMEFF2 | 0.0011092 | 834 | GTEx | DepMap | Descartes | 1.76 | 134.46 |
| CNKSR2 | 0.0008973 | 1066 | GTEx | DepMap | Descartes | 0.33 | 9.22 |
| PTCHD1 | 0.0008826 | 1092 | GTEx | DepMap | Descartes | 0.51 | 10.20 |
| TMEM132C | 0.0008165 | 1191 | GTEx | DepMap | Descartes | 0.25 | 14.81 |
| MARCH11 | 0.0006393 | 1553 | GTEx | DepMap | Descartes | 0.59 | NA |
| CNTFR | 0.0005724 | 1711 | GTEx | DepMap | Descartes | 1.46 | 188.51 |
| FAT3 | 0.0004184 | 2209 | GTEx | DepMap | Descartes | 0.15 | 2.42 |
| PLXNA4 | 0.0001980 | 3269 | GTEx | DepMap | Descartes | 0.53 | 8.78 |
| RGMB | 0.0001823 | 3367 | GTEx | DepMap | Descartes | 0.95 | 42.76 |
| ANKFN1 | 0.0001433 | 3629 | GTEx | DepMap | Descartes | 0.12 | 7.72 |
| ELAVL2 | 0.0001330 | 3700 | GTEx | DepMap | Descartes | 1.70 | 86.72 |
| MAP1B | 0.0000392 | 4514 | GTEx | DepMap | Descartes | 12.84 | 217.83 |
| KCNB2 | -0.0001140 | 6591 | GTEx | DepMap | Descartes | 0.07 | 4.05 |
| EPHA6 | -0.0001808 | 7667 | GTEx | DepMap | Descartes | 0.04 | 2.04 |
| SYNPO2 | -0.0002061 | 8023 | GTEx | DepMap | Descartes | 0.13 | 1.22 |
| RPH3A | -0.0002217 | 8256 | GTEx | DepMap | Descartes | 0.05 | 1.14 |
Descartes Vascular_endothelial markers
Top 50 marker genes of Vascular_endothelial cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/vascular_endothelial/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.94e-01
Mean rank of genes in gene set: 7735.05
Median rank of genes in gene set: 8376.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| IRX3 | 0.0007394 | 1316 | GTEx | DepMap | Descartes | 0.13 | 10.70 |
| RAMP2 | 0.0003541 | 2474 | GTEx | DepMap | Descartes | 1.48 | 402.57 |
| ARHGAP29 | 0.0001632 | 3505 | GTEx | DepMap | Descartes | 0.57 | 13.99 |
| EHD3 | 0.0000457 | 4460 | GTEx | DepMap | Descartes | 0.10 | 4.76 |
| CEACAM1 | -0.0000225 | 5164 | GTEx | DepMap | Descartes | 0.11 | 8.19 |
| PODXL | -0.0000439 | 5442 | GTEx | DepMap | Descartes | 0.10 | 5.24 |
| GALNT15 | -0.0000613 | 5732 | GTEx | DepMap | Descartes | 0.01 | NA |
| MYRIP | -0.0000772 | 5978 | GTEx | DepMap | Descartes | 0.04 | 0.89 |
| NR5A2 | -0.0001454 | 7110 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CYP26B1 | -0.0001524 | 7232 | GTEx | DepMap | Descartes | 0.01 | 0.11 |
| CHRM3 | -0.0001559 | 7296 | GTEx | DepMap | Descartes | 0.28 | 4.82 |
| SHE | -0.0001612 | 7391 | GTEx | DepMap | Descartes | 0.01 | 0.34 |
| ESM1 | -0.0001665 | 7478 | GTEx | DepMap | Descartes | 0.02 | 0.52 |
| CRHBP | -0.0001891 | 7776 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CDH13 | -0.0001943 | 7848 | GTEx | DepMap | Descartes | 0.04 | 0.63 |
| SHANK3 | -0.0001963 | 7866 | GTEx | DepMap | Descartes | 0.03 | 0.74 |
| BTNL9 | -0.0001995 | 7923 | GTEx | DepMap | Descartes | 0.03 | 2.14 |
| FLT4 | -0.0002005 | 7944 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| KDR | -0.0002224 | 8267 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TEK | -0.0002374 | 8486 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SLCO2A1 | -0.0002376 | 8495 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| KANK3 | -0.0002398 | 8524 | GTEx | DepMap | Descartes | 0.01 | 0.48 |
| NOTCH4 | -0.0002489 | 8631 | GTEx | DepMap | Descartes | 0.13 | 4.30 |
| F8 | -0.0002549 | 8714 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| NPR1 | -0.0002570 | 8746 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| HYAL2 | -0.0002652 | 8850 | GTEx | DepMap | Descartes | 0.29 | 15.02 |
| ROBO4 | -0.0002687 | 8898 | GTEx | DepMap | Descartes | 0.03 | 1.55 |
| MMRN2 | -0.0002744 | 8970 | GTEx | DepMap | Descartes | 0.01 | 0.10 |
| RASIP1 | -0.0002748 | 8974 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PTPRB | -0.0002767 | 8991 | GTEx | DepMap | Descartes | 0.02 | 0.24 |
Descartes stromal markers
Top 50 marker genes of stromal cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/stromal/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 8204.55
Median rank of genes in gene set: 8976
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| GAS2 | 0.0025010 | 233 | GTEx | DepMap | Descartes | 0.09 | 7.11 |
| PCOLCE | 0.0007205 | 1349 | GTEx | DepMap | Descartes | 1.46 | 249.94 |
| LAMC3 | 0.0000940 | 4024 | GTEx | DepMap | Descartes | 0.01 | 0.31 |
| COL27A1 | 0.0000203 | 4719 | GTEx | DepMap | Descartes | 0.02 | 0.66 |
| ADAMTS2 | -0.0000007 | 4949 | GTEx | DepMap | Descartes | 0.11 | 4.06 |
| GLI2 | -0.0000497 | 5535 | GTEx | DepMap | Descartes | 0.01 | 0.23 |
| FREM1 | -0.0000627 | 5755 | GTEx | DepMap | Descartes | 0.02 | 0.36 |
| PRICKLE1 | -0.0000874 | 6127 | GTEx | DepMap | Descartes | 0.13 | 4.50 |
| LRRC17 | -0.0000902 | 6173 | GTEx | DepMap | Descartes | 0.06 | 4.33 |
| ITGA11 | -0.0001233 | 6745 | GTEx | DepMap | Descartes | 0.05 | 1.16 |
| ADAMTSL3 | -0.0001320 | 6881 | GTEx | DepMap | Descartes | 0.03 | 0.99 |
| CLDN11 | -0.0001825 | 7694 | GTEx | DepMap | Descartes | 0.05 | 2.67 |
| RSPO3 | -0.0002056 | 8018 | GTEx | DepMap | Descartes | 0.00 | NA |
| DKK2 | -0.0002081 | 8060 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PAMR1 | -0.0002140 | 8145 | GTEx | DepMap | Descartes | 0.02 | 0.65 |
| SFRP2 | -0.0002171 | 8188 | GTEx | DepMap | Descartes | 0.01 | 0.12 |
| EDNRA | -0.0002295 | 8370 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SCARA5 | -0.0002319 | 8399 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| C7 | -0.0002649 | 8847 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PCDH18 | -0.0002659 | 8859 | GTEx | DepMap | Descartes | 0.01 | 0.74 |
| BICC1 | -0.0002714 | 8931 | GTEx | DepMap | Descartes | 0.03 | 1.65 |
| ABCC9 | -0.0002732 | 8951 | GTEx | DepMap | Descartes | 0.01 | 0.17 |
| COL6A3 | -0.0002776 | 9001 | GTEx | DepMap | Descartes | 0.15 | 3.04 |
| IGFBP3 | -0.0002912 | 9165 | GTEx | DepMap | Descartes | 0.06 | 5.24 |
| PDGFRA | -0.0003010 | 9264 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ACTA2 | -0.0003037 | 9291 | GTEx | DepMap | Descartes | 0.11 | 24.70 |
| ISLR | -0.0003041 | 9294 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PRRX1 | -0.0003045 | 9304 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ABCA6 | -0.0003214 | 9514 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| POSTN | -0.0003231 | 9536 | GTEx | DepMap | Descartes | 0.01 | 0.08 |
Descartes sympathoblasts markers
Top 50 marker genes of sympathoblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/sympathoblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 3.59e-03
Mean rank of genes in gene set: 4697.68
Median rank of genes in gene set: 4266
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| FGF14 | 0.0029090 | 165 | GTEx | DepMap | Descartes | 0.80 | 17.36 |
| NTNG1 | 0.0024592 | 245 | GTEx | DepMap | Descartes | 0.58 | 30.86 |
| KSR2 | 0.0023915 | 260 | GTEx | DepMap | Descartes | 0.20 | 3.35 |
| GALNTL6 | 0.0022800 | 282 | GTEx | DepMap | Descartes | 0.25 | 19.64 |
| CHGA | 0.0022098 | 300 | GTEx | DepMap | Descartes | 3.67 | 456.03 |
| CCSER1 | 0.0018801 | 405 | GTEx | DepMap | Descartes | 0.35 | NA |
| PCSK2 | 0.0010634 | 880 | GTEx | DepMap | Descartes | 0.16 | 9.37 |
| TENM1 | 0.0009542 | 1002 | GTEx | DepMap | Descartes | 0.29 | NA |
| UNC80 | 0.0008710 | 1109 | GTEx | DepMap | Descartes | 0.54 | 9.86 |
| TIAM1 | 0.0008347 | 1162 | GTEx | DepMap | Descartes | 0.46 | 15.34 |
| KCTD16 | 0.0008167 | 1189 | GTEx | DepMap | Descartes | 0.78 | 15.29 |
| EML6 | 0.0007201 | 1352 | GTEx | DepMap | Descartes | 0.21 | 5.10 |
| SPOCK3 | 0.0005600 | 1748 | GTEx | DepMap | Descartes | 0.58 | 44.31 |
| CDH18 | 0.0005165 | 1873 | GTEx | DepMap | Descartes | 0.15 | 7.71 |
| MGAT4C | 0.0004450 | 2115 | GTEx | DepMap | Descartes | 0.30 | 2.31 |
| CDH12 | 0.0002063 | 3215 | GTEx | DepMap | Descartes | 0.12 | 5.44 |
| FAM155A | 0.0001436 | 3628 | GTEx | DepMap | Descartes | 0.26 | 5.70 |
| TBX20 | 0.0000789 | 4167 | GTEx | DepMap | Descartes | 0.03 | 5.50 |
| DGKK | 0.0000684 | 4249 | GTEx | DepMap | Descartes | 0.07 | 3.23 |
| GRID2 | 0.0000652 | 4283 | GTEx | DepMap | Descartes | 0.05 | 1.89 |
| CHGB | -0.0000155 | 5101 | GTEx | DepMap | Descartes | 4.76 | 460.31 |
| SLC35F3 | -0.0000265 | 5214 | GTEx | DepMap | Descartes | 0.04 | 2.44 |
| ARC | -0.0000369 | 5345 | GTEx | DepMap | Descartes | 0.31 | 26.88 |
| AGBL4 | -0.0000579 | 5666 | GTEx | DepMap | Descartes | 0.02 | 1.49 |
| ST18 | -0.0000957 | 6262 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| LAMA3 | -0.0001388 | 6996 | GTEx | DepMap | Descartes | 0.03 | 1.15 |
| PCSK1N | -0.0001395 | 7004 | GTEx | DepMap | Descartes | 5.47 | 1109.07 |
| PENK | -0.0001482 | 7157 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SLC24A2 | -0.0001493 | 7171 | GTEx | DepMap | Descartes | 0.01 | 0.23 |
| GRM7 | -0.0001635 | 7430 | GTEx | DepMap | Descartes | 0.02 | 0.46 |
Descartes erythroblasts markers
Top 50 marker genes of erythroblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/erythroblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 7.92e-01
Mean rank of genes in gene set: 6822.55
Median rank of genes in gene set: 6633
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TFR2 | 0.0024473 | 250 | GTEx | DepMap | Descartes | 0.66 | 51.77 |
| SOX6 | 0.0008311 | 1173 | GTEx | DepMap | Descartes | 0.29 | 9.47 |
| TSPAN5 | 0.0007640 | 1271 | GTEx | DepMap | Descartes | 0.97 | 55.81 |
| XPO7 | 0.0002403 | 3003 | GTEx | DepMap | Descartes | 0.41 | 19.08 |
| SPTB | 0.0001591 | 3531 | GTEx | DepMap | Descartes | 0.08 | 2.14 |
| DENND4A | 0.0000458 | 4458 | GTEx | DepMap | Descartes | 0.49 | 16.08 |
| FECH | 0.0000232 | 4688 | GTEx | DepMap | Descartes | 0.31 | 8.64 |
| GCLC | -0.0000176 | 5121 | GTEx | DepMap | Descartes | 0.23 | 13.68 |
| RHD | -0.0000373 | 5353 | GTEx | DepMap | Descartes | 0.02 | 1.24 |
| SLC25A37 | -0.0000485 | 5513 | GTEx | DepMap | Descartes | 0.93 | 53.77 |
| SLC25A21 | -0.0000689 | 5857 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SNCA | -0.0000711 | 5891 | GTEx | DepMap | Descartes | 0.76 | 45.83 |
| ALAS2 | -0.0001114 | 6548 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ABCB10 | -0.0001125 | 6564 | GTEx | DepMap | Descartes | 0.11 | 7.97 |
| RGS6 | -0.0001166 | 6633 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SLC4A1 | -0.0001402 | 7016 | GTEx | DepMap | Descartes | 0.01 | 0.63 |
| ANK1 | -0.0001548 | 7272 | GTEx | DepMap | Descartes | 0.10 | 2.88 |
| RAPGEF2 | -0.0001840 | 7713 | GTEx | DepMap | Descartes | 0.21 | 4.95 |
| SELENBP1 | -0.0002430 | 8559 | GTEx | DepMap | Descartes | 0.01 | 0.33 |
| TMCC2 | -0.0002476 | 8616 | GTEx | DepMap | Descartes | 0.03 | 1.82 |
| SPECC1 | -0.0002745 | 8971 | GTEx | DepMap | Descartes | 0.07 | 1.94 |
| MICAL2 | -0.0002794 | 9017 | GTEx | DepMap | Descartes | 0.03 | 0.72 |
| CPOX | -0.0002966 | 9221 | GTEx | DepMap | Descartes | 0.06 | 3.54 |
| TRAK2 | -0.0003274 | 9583 | GTEx | DepMap | Descartes | 0.16 | 4.64 |
| MARCH3 | -0.0003300 | 9614 | GTEx | DepMap | Descartes | 0.04 | NA |
| CAT | -0.0004631 | 10709 | GTEx | DepMap | Descartes | 0.45 | 58.23 |
| EPB41 | -0.0005858 | 11365 | GTEx | DepMap | Descartes | 0.20 | 7.24 |
| BLVRB | -0.0007640 | 11897 | GTEx | DepMap | Descartes | 0.27 | 22.08 |
| GYPC | -0.0013452 | 12447 | GTEx | DepMap | Descartes | 0.06 | 9.34 |
| NA | NA | NA | NA | NA | NA | NA | NA |
Descartes myeloid markers
Top 50 marker genes of myeloid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/myeloid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 10318.26
Median rank of genes in gene set: 11109
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CD163L1 | 0.0025308 | 226 | GTEx | DepMap | Descartes | 2.01 | 104.37 |
| RBPJ | 0.0002874 | 2755 | GTEx | DepMap | Descartes | 1.71 | 71.84 |
| HRH1 | 0.0000164 | 4772 | GTEx | DepMap | Descartes | 0.03 | 1.99 |
| RGL1 | -0.0001673 | 7490 | GTEx | DepMap | Descartes | 0.07 | 3.23 |
| ATP8B4 | -0.0002392 | 8515 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MSR1 | -0.0002763 | 8988 | GTEx | DepMap | Descartes | 0.02 | 0.57 |
| SPP1 | -0.0002916 | 9168 | GTEx | DepMap | Descartes | 0.41 | 40.56 |
| SLC1A3 | -0.0003149 | 9426 | GTEx | DepMap | Descartes | 0.02 | 0.25 |
| MERTK | -0.0003155 | 9434 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SLCO2B1 | -0.0003370 | 9696 | GTEx | DepMap | Descartes | 0.02 | 0.36 |
| ABCA1 | -0.0003669 | 9994 | GTEx | DepMap | Descartes | 0.12 | 1.84 |
| WWP1 | -0.0003996 | 10255 | GTEx | DepMap | Descartes | 0.07 | 2.46 |
| SLC9A9 | -0.0004372 | 10554 | GTEx | DepMap | Descartes | 0.02 | 1.10 |
| ADAP2 | -0.0004400 | 10570 | GTEx | DepMap | Descartes | 0.01 | 0.98 |
| LGMN | -0.0004689 | 10743 | GTEx | DepMap | Descartes | 0.31 | 19.83 |
| SFMBT2 | -0.0004703 | 10755 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MS4A4A | -0.0004738 | 10778 | GTEx | DepMap | Descartes | 0.02 | 0.45 |
| AXL | -0.0004826 | 10831 | GTEx | DepMap | Descartes | 0.07 | 1.49 |
| CD163 | -0.0005111 | 10982 | GTEx | DepMap | Descartes | 0.01 | 0.47 |
| FMN1 | -0.0005583 | 11236 | GTEx | DepMap | Descartes | 0.08 | 0.86 |
| FGD2 | -0.0005594 | 11243 | GTEx | DepMap | Descartes | 0.08 | 2.84 |
| CSF1R | -0.0006002 | 11407 | GTEx | DepMap | Descartes | 0.02 | 0.25 |
| ITPR2 | -0.0006010 | 11409 | GTEx | DepMap | Descartes | 0.04 | 0.49 |
| CTSD | -0.0006665 | 11651 | GTEx | DepMap | Descartes | 1.26 | 112.50 |
| TGFBI | -0.0006782 | 11687 | GTEx | DepMap | Descartes | 0.01 | 0.23 |
| MARCH1 | -0.0006792 | 11690 | GTEx | DepMap | Descartes | 0.04 | NA |
| CD14 | -0.0006920 | 11731 | GTEx | DepMap | Descartes | 0.05 | 7.25 |
| CPVL | -0.0007740 | 11916 | GTEx | DepMap | Descartes | 0.05 | 3.06 |
| CYBB | -0.0007965 | 11966 | GTEx | DepMap | Descartes | 0.09 | 3.26 |
| FGL2 | -0.0008188 | 12007 | GTEx | DepMap | Descartes | 0.08 | 2.34 |
Descartes Schwann markers
Top 50 marker genes of Schwann cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/schwann/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.17e-01
Mean rank of genes in gene set: 7029.82
Median rank of genes in gene set: 6742
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ERBB4 | 0.0039742 | 74 | GTEx | DepMap | Descartes | 0.79 | 16.94 |
| MARCKS | 0.0027086 | 191 | GTEx | DepMap | Descartes | 12.73 | 731.73 |
| LAMB1 | 0.0018501 | 420 | GTEx | DepMap | Descartes | 1.02 | 48.39 |
| DST | 0.0010189 | 929 | GTEx | DepMap | Descartes | 2.94 | 35.67 |
| FIGN | 0.0004725 | 2016 | GTEx | DepMap | Descartes | 0.24 | 6.95 |
| SOX5 | 0.0002998 | 2690 | GTEx | DepMap | Descartes | 0.12 | 3.15 |
| GFRA3 | 0.0002356 | 3037 | GTEx | DepMap | Descartes | 1.26 | 146.36 |
| PLCE1 | 0.0002207 | 3125 | GTEx | DepMap | Descartes | 0.20 | 3.98 |
| SLC35F1 | 0.0001927 | 3300 | GTEx | DepMap | Descartes | 0.13 | 5.86 |
| SFRP1 | 0.0001722 | 3441 | GTEx | DepMap | Descartes | 1.28 | 63.38 |
| ERBB3 | 0.0001685 | 3466 | GTEx | DepMap | Descartes | 0.02 | 0.64 |
| NRXN3 | 0.0000497 | 4426 | GTEx | DepMap | Descartes | 0.02 | 0.34 |
| IL1RAPL2 | 0.0000227 | 4694 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| EGFLAM | -0.0000377 | 5358 | GTEx | DepMap | Descartes | 0.04 | 1.25 |
| COL25A1 | -0.0000646 | 5789 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PTPRZ1 | -0.0000723 | 5916 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MDGA2 | -0.0000940 | 6228 | GTEx | DepMap | Descartes | 0.02 | 0.19 |
| STARD13 | -0.0001036 | 6400 | GTEx | DepMap | Descartes | 0.01 | 0.14 |
| HMGA2 | -0.0001038 | 6405 | GTEx | DepMap | Descartes | 0.01 | 0.30 |
| GRIK3 | -0.0001112 | 6543 | GTEx | DepMap | Descartes | 0.02 | 0.58 |
| XKR4 | -0.0001123 | 6558 | GTEx | DepMap | Descartes | 0.09 | 1.40 |
| TRPM3 | -0.0001142 | 6593 | GTEx | DepMap | Descartes | 0.13 | 1.69 |
| MPZ | -0.0001328 | 6891 | GTEx | DepMap | Descartes | 0.02 | 1.50 |
| PLP1 | -0.0002135 | 8135 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| LRRTM4 | -0.0002463 | 8599 | GTEx | DepMap | Descartes | 0.14 | 6.19 |
| IL1RAPL1 | -0.0002519 | 8674 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ADAMTS5 | -0.0002589 | 8772 | GTEx | DepMap | Descartes | 0.01 | 0.19 |
| NRXN1 | -0.0002774 | 8998 | GTEx | DepMap | Descartes | 0.55 | 8.74 |
| OLFML2A | -0.0002840 | 9086 | GTEx | DepMap | Descartes | 0.01 | 0.77 |
| SORCS1 | -0.0002966 | 9222 | GTEx | DepMap | Descartes | 0.08 | 1.66 |
Descartes Megakaryocytes markers
Top 50 marker genes of Megakaryocytes cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/megakaryocytes/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 9312.71
Median rank of genes in gene set: 10620
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| PDE3A | 0.0010108 | 935 | GTEx | DepMap | Descartes | 0.13 | 4.05 |
| RAB27B | 0.0006617 | 1490 | GTEx | DepMap | Descartes | 0.35 | 11.06 |
| MED12L | 0.0005895 | 1667 | GTEx | DepMap | Descartes | 0.05 | 1.26 |
| HIPK2 | 0.0004723 | 2017 | GTEx | DepMap | Descartes | 0.90 | 13.93 |
| ITGA2B | 0.0000366 | 4539 | GTEx | DepMap | Descartes | 0.04 | 2.65 |
| GP1BA | -0.0000045 | 4992 | GTEx | DepMap | Descartes | 0.03 | 2.38 |
| DOK6 | -0.0000364 | 5339 | GTEx | DepMap | Descartes | 0.12 | 2.58 |
| TUBB1 | -0.0000482 | 5512 | GTEx | DepMap | Descartes | 0.03 | 2.14 |
| SLC24A3 | -0.0001208 | 6711 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ITGB3 | -0.0001404 | 7021 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ANGPT1 | -0.0001434 | 7069 | GTEx | DepMap | Descartes | 0.01 | 0.58 |
| TRPC6 | -0.0001744 | 7586 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ARHGAP6 | -0.0001787 | 7636 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PRKAR2B | -0.0002200 | 8230 | GTEx | DepMap | Descartes | 0.56 | 30.85 |
| MMRN1 | -0.0002451 | 8588 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| STON2 | -0.0002619 | 8803 | GTEx | DepMap | Descartes | 0.08 | 2.88 |
| MYLK | -0.0002811 | 9045 | GTEx | DepMap | Descartes | 0.03 | 0.56 |
| LTBP1 | -0.0003203 | 9501 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| INPP4B | -0.0003678 | 10001 | GTEx | DepMap | Descartes | 0.03 | 0.64 |
| P2RX1 | -0.0003898 | 10175 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| VCL | -0.0004203 | 10424 | GTEx | DepMap | Descartes | 0.14 | 4.30 |
| MCTP1 | -0.0004257 | 10475 | GTEx | DepMap | Descartes | 0.01 | 0.19 |
| GSN | -0.0004473 | 10620 | GTEx | DepMap | Descartes | 0.08 | 2.29 |
| PSTPIP2 | -0.0004588 | 10683 | GTEx | DepMap | Descartes | 0.03 | 0.67 |
| STOM | -0.0004840 | 10840 | GTEx | DepMap | Descartes | 0.30 | 20.81 |
| CD84 | -0.0004907 | 10865 | GTEx | DepMap | Descartes | 0.13 | 1.49 |
| UBASH3B | -0.0004933 | 10876 | GTEx | DepMap | Descartes | 0.04 | 1.25 |
| ZYX | -0.0005010 | 10925 | GTEx | DepMap | Descartes | 0.64 | 73.39 |
| ACTN1 | -0.0005513 | 11201 | GTEx | DepMap | Descartes | 0.34 | 14.07 |
| LIMS1 | -0.0005878 | 11371 | GTEx | DepMap | Descartes | 0.86 | 38.72 |
Descartes Lyphoid markers
Top 50 marker genes of Lyphoid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/lymphoid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 10854.69
Median rank of genes in gene set: 11828
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| SCML4 | 0.0002564 | 2925 | GTEx | DepMap | Descartes | 0.70 | 40.19 |
| STK39 | 0.0001147 | 3852 | GTEx | DepMap | Descartes | 0.51 | 30.75 |
| BCL2 | 0.0000519 | 4406 | GTEx | DepMap | Descartes | 0.50 | 15.92 |
| SORL1 | -0.0001295 | 6839 | GTEx | DepMap | Descartes | 0.67 | 15.87 |
| ABLIM1 | -0.0001896 | 7783 | GTEx | DepMap | Descartes | 0.38 | 11.36 |
| EVL | -0.0002636 | 8832 | GTEx | DepMap | Descartes | 2.91 | 186.81 |
| PDE3B | -0.0003039 | 9292 | GTEx | DepMap | Descartes | 0.38 | 19.04 |
| FYN | -0.0003607 | 9943 | GTEx | DepMap | Descartes | 2.02 | 125.23 |
| RAP1GAP2 | -0.0003611 | 9946 | GTEx | DepMap | Descartes | 0.16 | 4.59 |
| ITPKB | -0.0003825 | 10111 | GTEx | DepMap | Descartes | 0.04 | 1.86 |
| FOXP1 | -0.0004580 | 10678 | GTEx | DepMap | Descartes | 1.88 | 48.26 |
| PITPNC1 | -0.0004686 | 10740 | GTEx | DepMap | Descartes | 0.19 | 6.32 |
| TOX | -0.0004783 | 10802 | GTEx | DepMap | Descartes | 0.08 | 4.23 |
| SAMD3 | -0.0005269 | 11062 | GTEx | DepMap | Descartes | 0.02 | 1.74 |
| BACH2 | -0.0005449 | 11159 | GTEx | DepMap | Descartes | 0.31 | 6.95 |
| GNG2 | -0.0005808 | 11337 | GTEx | DepMap | Descartes | 0.98 | 66.12 |
| NCALD | -0.0006060 | 11437 | GTEx | DepMap | Descartes | 0.06 | 2.05 |
| DOCK10 | -0.0006159 | 11475 | GTEx | DepMap | Descartes | 0.09 | 2.30 |
| LEF1 | -0.0006740 | 11678 | GTEx | DepMap | Descartes | 0.01 | 0.19 |
| SKAP1 | -0.0006825 | 11701 | GTEx | DepMap | Descartes | 0.03 | 3.81 |
| MCTP2 | -0.0007001 | 11756 | GTEx | DepMap | Descartes | 0.01 | 0.10 |
| PLEKHA2 | -0.0007653 | 11900 | GTEx | DepMap | Descartes | 0.06 | 2.05 |
| ANKRD44 | -0.0007794 | 11928 | GTEx | DepMap | Descartes | 0.23 | 7.62 |
| CCL5 | -0.0008030 | 11980 | GTEx | DepMap | Descartes | 0.72 | 118.74 |
| PRKCH | -0.0008061 | 11984 | GTEx | DepMap | Descartes | 0.02 | 1.66 |
| ARHGAP15 | -0.0008949 | 12118 | GTEx | DepMap | Descartes | 0.04 | 2.70 |
| ARID5B | -0.0009314 | 12164 | GTEx | DepMap | Descartes | 0.05 | 0.98 |
| IKZF1 | -0.0009466 | 12189 | GTEx | DepMap | Descartes | 0.01 | 0.38 |
| MBNL1 | -0.0009694 | 12216 | GTEx | DepMap | Descartes | 0.48 | 16.74 |
| ETS1 | -0.0010608 | 12296 | GTEx | DepMap | Descartes | 0.02 | 1.03 |
Below shows the ranks on this GEP for immune marker gene lists curated by CellTypist (https://www.celltypist.org/encyclopedia).
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Erythroid: Mid erythroid (curated markers)
middle erythroid cells in fetal liver:
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.20e-02
Mean rank of genes in gene set: 497
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| PRDX2 | 0.0016871 | 482 | GTEx | DepMap | Descartes | 7.57 | 1065.99 |
| KCNH2 | 0.0016242 | 512 | GTEx | DepMap | Descartes | 0.83 | 41.15 |
pDC: pDC (model markers)
rare plasmacytoid dendritic cell subpopulation which serves as the major source of type I interferons when the body is infected by a virus:
Wilcoxon ranksum test P-value for gene set overrepresentation: 3.66e-02
Mean rank of genes in gene set: 1685.5
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TRIM71 | 0.0008019 | 1213 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| OGDHL | 0.0004318 | 2158 | GTEx | DepMap | Descartes | 0.07 | 4.51 |
No detectable expression in this dataset: OR2A25
HSC/MPP: Megakaryocyte-erythroid-mast cell progenitor (model markers)
shared progenitors in the fetal liver which originate from common myeloid progenitors and differentiate into megakaryocytes:
Wilcoxon ranksum test P-value for gene set overrepresentation: 5.03e-02
Mean rank of genes in gene set: 327
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CKB | 0.0021025 | 327 | GTEx | DepMap | Descartes | 4.85 | 753.49 |
No detectable expression in this dataset: OR2A25