Program: 41. IFN Response: Activity Program.
Submit a comment on this gene expression program’s interpretation: CLICK
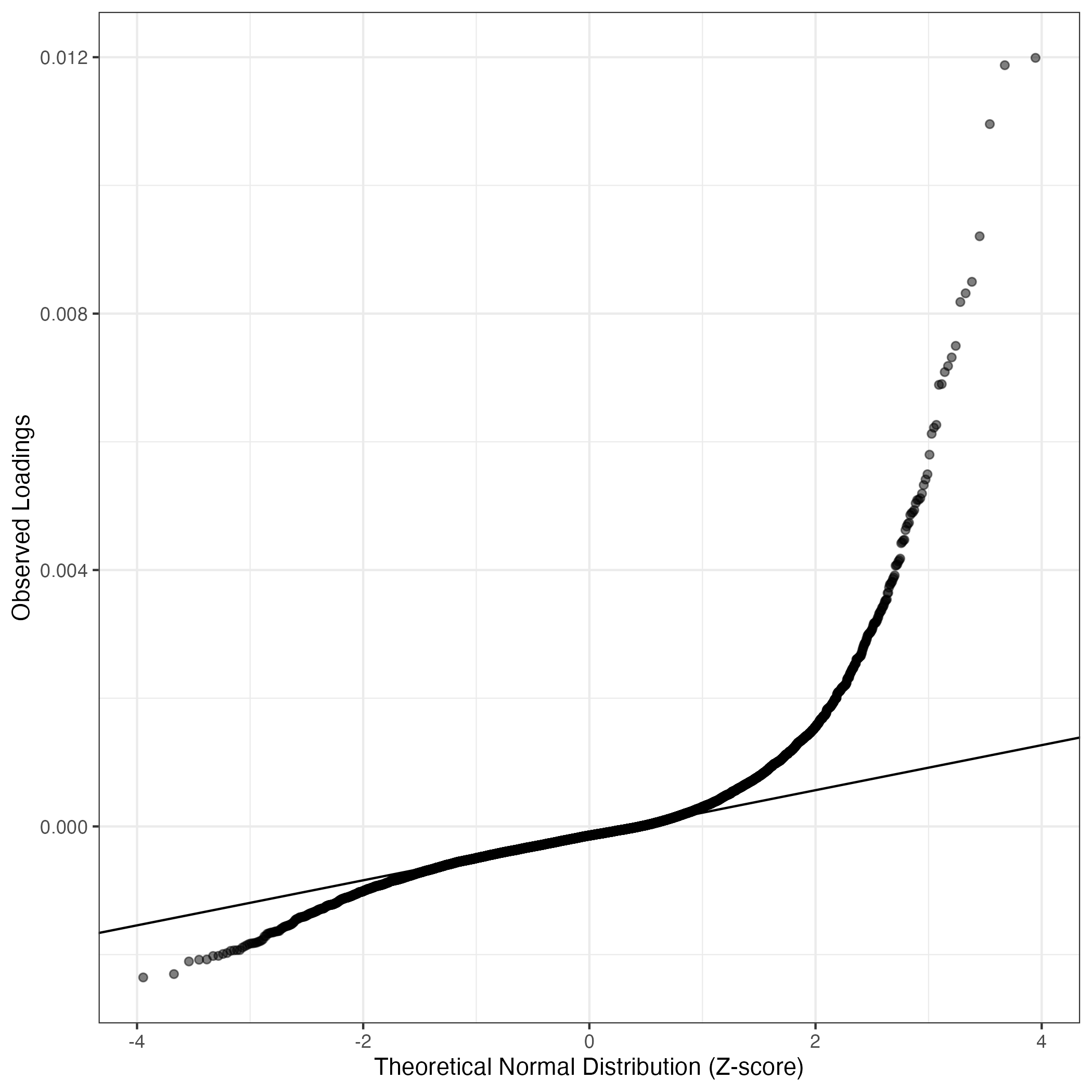
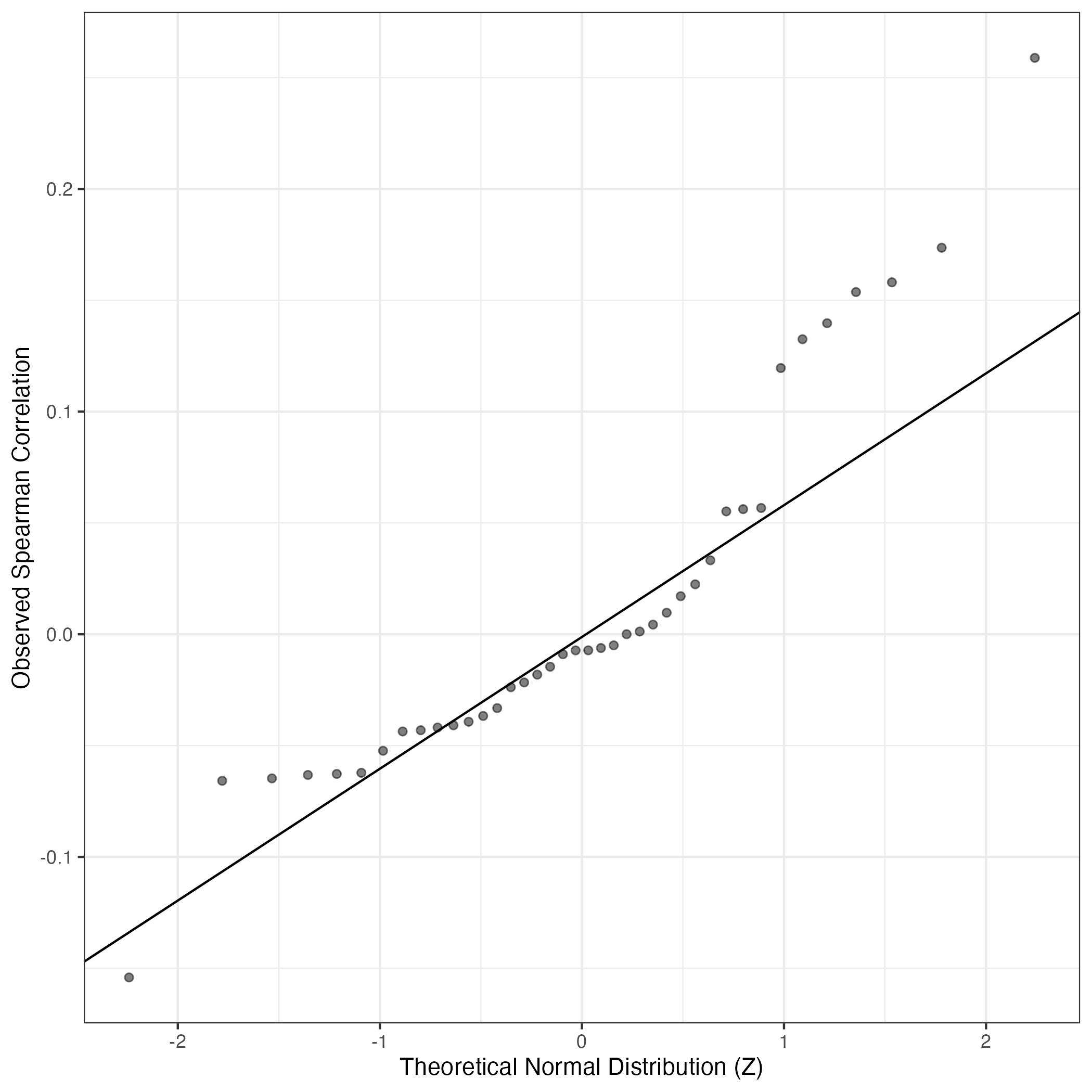
QQ-plot of gene loadings, averaged over both independent splits of the data
This plot highlights the relative contribution of each gene to the GEP
Top genes driving this program.
Note: Decartes website is buggy, try refreshing. Also, Decartes fetal adrenal data have been collected at specific time points (89-122 days), all possible cell types of interest may not be represented, do not overinterpret.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
| Gene | Loading | Gene.Name | GTEx | DepMap | Descartes | Mean.Counts | Mean.Tpm | |
|---|---|---|---|---|---|---|---|---|
| 1 | IFIT1 | 0.0119898 | interferon induced protein with tetratricopeptide repeats 1 | GTEx | DepMap | Descartes | 3.74 | 125.06 |
| 2 | IFI44 | 0.0118752 | interferon induced protein 44 | GTEx | DepMap | Descartes | 2.69 | 240.25 |
| 3 | IFIT3 | 0.0109563 | interferon induced protein with tetratricopeptide repeats 3 | GTEx | DepMap | Descartes | 2.88 | 178.74 |
| 4 | IFI27 | 0.0092068 | interferon alpha inducible protein 27 | GTEx | DepMap | Descartes | 26.52 | 1606.78 |
| 5 | CRABP1 | 0.0084962 | cellular retinoic acid binding protein 1 | GTEx | DepMap | Descartes | 2.28 | 434.81 |
| 6 | STAT1 | 0.0083173 | signal transducer and activator of transcription 1 | GTEx | DepMap | Descartes | 3.98 | 141.58 |
| 7 | NOS1 | 0.0081810 | nitric oxide synthase 1 | GTEx | DepMap | Descartes | 0.35 | 4.13 |
| 8 | SPATS2L | 0.0074968 | spermatogenesis associated serine rich 2 like | GTEx | DepMap | Descartes | 3.55 | 89.39 |
| 9 | FGF10 | 0.0073169 | fibroblast growth factor 10 | GTEx | DepMap | Descartes | 0.49 | 12.14 |
| 10 | EIF2AK2 | 0.0071824 | eukaryotic translation initiation factor 2 alpha kinase 2 | GTEx | DepMap | Descartes | 3.89 | 57.87 |
| 11 | CALM1 | 0.0070880 | calmodulin 1 | GTEx | DepMap | Descartes | 25.25 | 900.56 |
| 12 | CNR1 | 0.0069005 | cannabinoid receptor 1 | GTEx | DepMap | Descartes | 0.83 | 19.91 |
| 13 | NTS | 0.0068891 | neurotensin | GTEx | DepMap | Descartes | 0.75 | 102.25 |
| 14 | FBXO8 | 0.0062642 | F-box protein 8 | GTEx | DepMap | Descartes | 1.39 | 106.09 |
| 15 | NPY | 0.0062200 | neuropeptide Y | GTEx | DepMap | Descartes | 54.84 | 11736.49 |
| 16 | UCHL1 | 0.0061251 | ubiquitin C-terminal hydrolase L1 | GTEx | DepMap | Descartes | 21.90 | 2050.19 |
| 17 | CSGALNACT1 | 0.0057991 | chondroitin sulfate N-acetylgalactosaminyltransferase 1 | GTEx | DepMap | Descartes | 1.87 | 69.59 |
| 18 | GREM2 | 0.0054936 | gremlin 2, DAN family BMP antagonist | GTEx | DepMap | Descartes | 0.34 | 11.79 |
| 19 | NBAS | 0.0054123 | NBAS subunit of NRZ tethering complex | GTEx | DepMap | Descartes | 19.89 | 354.87 |
| 20 | HGF | 0.0053251 | hepatocyte growth factor | GTEx | DepMap | Descartes | 2.64 | 67.00 |
| 21 | PKIB | 0.0051928 | cAMP-dependent protein kinase inhibitor beta | GTEx | DepMap | Descartes | 3.46 | 264.02 |
| 22 | NNAT | 0.0051197 | neuronatin | GTEx | DepMap | Descartes | 7.58 | 911.70 |
| 23 | RTN1 | 0.0050951 | reticulon 1 | GTEx | DepMap | Descartes | 16.39 | 763.13 |
| 24 | NRXN1 | 0.0050922 | neurexin 1 | GTEx | DepMap | Descartes | 3.83 | 64.45 |
| 25 | PLCB1 | 0.0050420 | phospholipase C beta 1 | GTEx | DepMap | Descartes | 0.87 | 17.74 |
| 26 | NR2F1 | 0.0049347 | nuclear receptor subfamily 2 group F member 1 | GTEx | DepMap | Descartes | 4.42 | 177.72 |
| 27 | MGAT4C | 0.0049011 | MGAT4 family member C | GTEx | DepMap | Descartes | 0.90 | 5.63 |
| 28 | ARX | 0.0048894 | aristaless related homeobox | GTEx | DepMap | Descartes | 0.71 | 38.54 |
| 29 | MAB21L2 | 0.0048588 | mab-21 like 2 | GTEx | DepMap | Descartes | 3.11 | 188.54 |
| 30 | EEF1A1 | 0.0047375 | eukaryotic translation elongation factor 1 alpha 1 | GTEx | DepMap | Descartes | 155.85 | 5097.37 |
| 31 | AIG1 | 0.0047192 | androgen induced 1 | GTEx | DepMap | Descartes | 2.54 | 104.34 |
| 32 | LRRN3 | 0.0046803 | leucine rich repeat neuronal 3 | GTEx | DepMap | Descartes | 4.53 | 184.21 |
| 33 | PMAIP1 | 0.0046232 | phorbol-12-myristate-13-acetate-induced protein 1 | GTEx | DepMap | Descartes | 1.91 | 153.06 |
| 34 | THADA | 0.0044712 | THADA armadillo repeat containing | GTEx | DepMap | Descartes | 1.92 | 41.81 |
| 35 | CTHRC1 | 0.0044624 | collagen triple helix repeat containing 1 | GTEx | DepMap | Descartes | 2.49 | 293.76 |
| 36 | GSG1L | 0.0044357 | GSG1 like | GTEx | DepMap | Descartes | 0.18 | 5.07 |
| 37 | PITX1 | 0.0044185 | paired like homeodomain 1 | GTEx | DepMap | Descartes | 0.19 | 12.31 |
| 38 | HMCN1 | 0.0041778 | hemicentin 1 | GTEx | DepMap | Descartes | 0.08 | 0.69 |
| 39 | TMEM108 | 0.0041530 | transmembrane protein 108 | GTEx | DepMap | Descartes | 0.83 | 25.59 |
| 40 | OAS3 | 0.0041313 | 2’-5’-oligoadenylate synthetase 3 | GTEx | DepMap | Descartes | 0.55 | 12.63 |
| 41 | SNCG | 0.0040906 | synuclein gamma | GTEx | DepMap | Descartes | 0.66 | 105.85 |
| 42 | NAP1L5 | 0.0040741 | nucleosome assembly protein 1 like 5 | GTEx | DepMap | Descartes | 1.66 | 135.46 |
| 43 | HERC6 | 0.0040685 | HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 | GTEx | DepMap | Descartes | 0.28 | 11.25 |
| 44 | FAM49A | 0.0039161 | NA | GTEx | DepMap | Descartes | 11.62 | NA |
| 45 | PCDHB6 | 0.0038871 | protocadherin beta 6 | GTEx | DepMap | Descartes | 0.66 | 30.14 |
| 46 | LGALS3BP | 0.0038619 | galectin 3 binding protein | GTEx | DepMap | Descartes | 2.98 | 199.68 |
| 47 | MMD | 0.0038208 | monocyte to macrophage differentiation associated | GTEx | DepMap | Descartes | 2.45 | 140.40 |
| 48 | CUTA | 0.0038037 | cutA divalent cation tolerance homolog | GTEx | DepMap | Descartes | 9.49 | 992.53 |
| 49 | ADM | 0.0037772 | adrenomedullin | GTEx | DepMap | Descartes | 1.05 | 88.45 |
| 50 | RTN4 | 0.0037733 | reticulon 4 | GTEx | DepMap | Descartes | 8.20 | 253.10 |
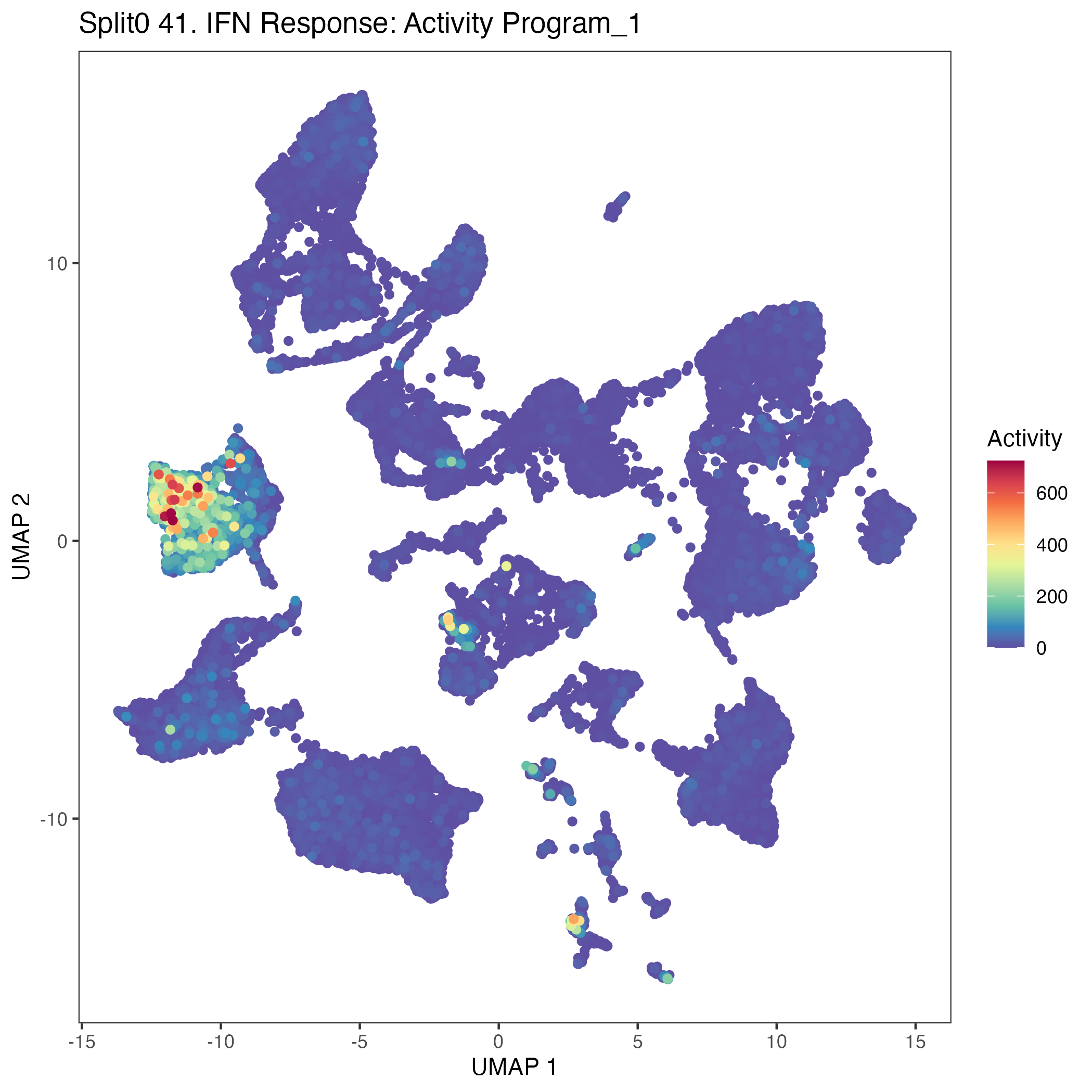
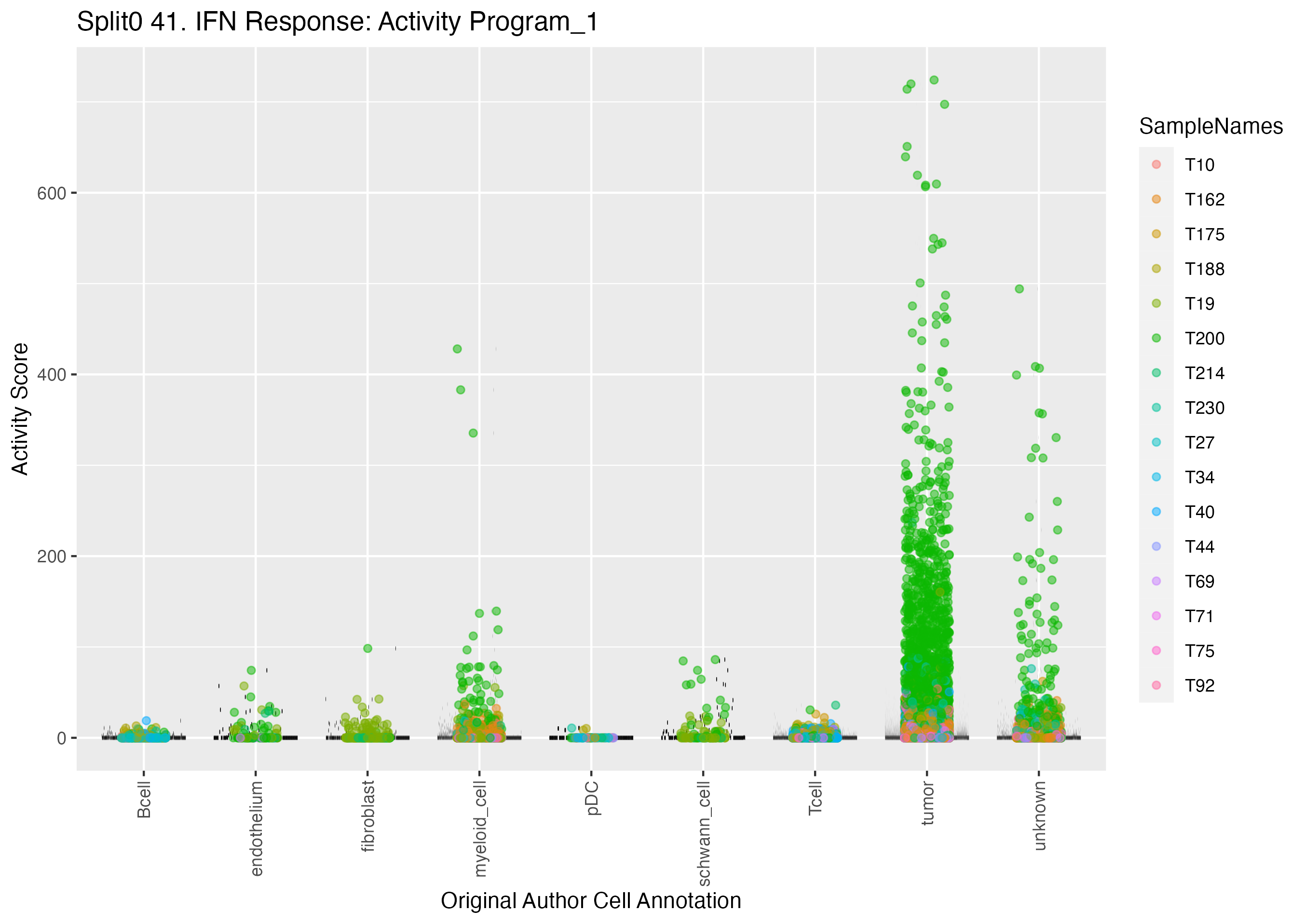
UMAP plots showing activity of gene expression program identified in GEP 41. IFN Response: Activity Program:
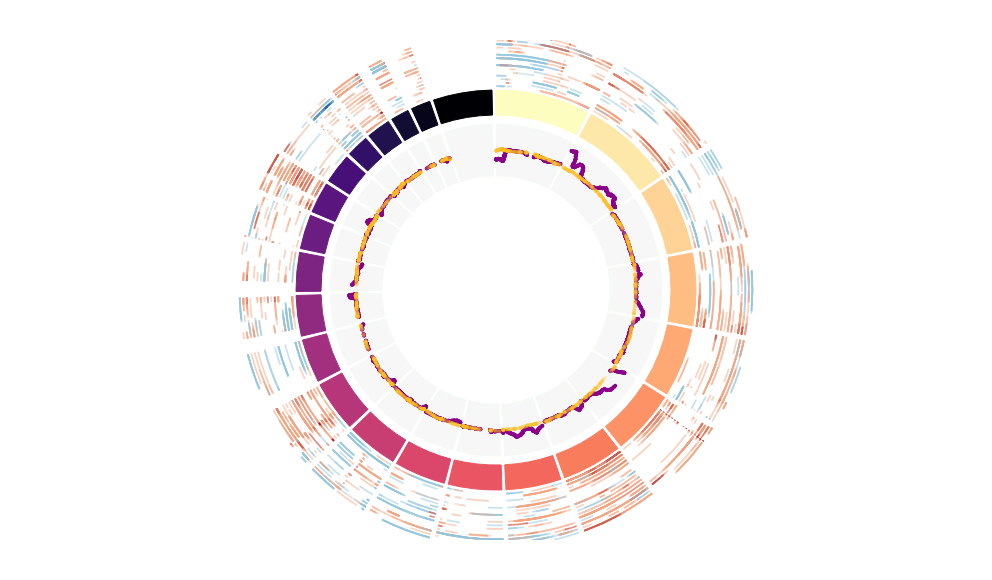
CNV Data procured from inferCNV.
Outer tracks are putative CNV regions (gains = red, losses = blue) for each patient
Inner track is expression data representing:
The top cells expressing this GEP (purple)
Random cells (n =50) from the reference set used in inferCNV (orange)
Gene set Enrichments for this program, caculated from top 50 genes
mSigDB Cell Types Gene Set:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| HU_FETAL_RETINA_AMACRINE | 1.17e-04 | 18.10 | 4.59 | 1.30e-02 | 7.82e-02 | 4CRABP1, NNAT, RTN1, NRXN1 |
64 |
| DESCARTES_FETAL_PANCREAS_ENS_NEURONS | 4.04e-05 | 10.85 | 3.73 | 1.04e-02 | 2.71e-02 | 6CRABP1, NOS1, CNR1, NPY, UCHL1, NNAT |
163 |
| TRAVAGLINI_LUNG_NEUROENDOCRINE_CELL | 1.43e-04 | 11.29 | 3.44 | 1.37e-02 | 9.59e-02 | 5UCHL1, PKIB, NNAT, NRXN1, PITX1 |
128 |
| DURANTE_ADULT_OLFACTORY_NEUROEPITHELIUM_MATURE_NEURONS | 8.59e-05 | 9.42 | 3.24 | 1.30e-02 | 5.76e-02 | 6IFI27, CALM1, UCHL1, RTN1, NRXN1, CUTA |
187 |
| MANNO_MIDBRAIN_NEUROTYPES_HRN | 3.98e-05 | 7.28 | 2.93 | 1.04e-02 | 2.67e-02 | 8CNR1, UCHL1, GREM2, NNAT, RTN1, NRXN1, SNCG, NAP1L5 |
335 |
| DESCARTES_FETAL_MUSCLE_SATELLITE_CELLS | 1.55e-03 | 14.50 | 2.80 | 8.69e-02 | 1.00e+00 | 3SPATS2L, NPY, GSG1L |
58 |
| MANNO_MIDBRAIN_NEUROTYPES_HSERT | 4.67e-05 | 6.22 | 2.64 | 1.04e-02 | 3.13e-02 | 9UCHL1, GREM2, RTN1, NRXN1, PLCB1, MGAT4C, CTHRC1, SNCG, NAP1L5 |
450 |
| DESCARTES_MAIN_FETAL_CHROMAFFIN_CELLS | 4.05e-03 | 23.65 | 2.62 | 1.60e-01 | 1.00e+00 | 2NPY, MAB21L2 |
24 |
| MENON_FETAL_KIDNEY_5_PROXIMAL_TUBULE_CELLS | 9.77e-04 | 10.06 | 2.58 | 8.03e-02 | 6.56e-01 | 4SPATS2L, CALM1, AIG1, RTN4 |
112 |
| MANNO_MIDBRAIN_NEUROTYPES_HOMTN | 1.10e-04 | 6.25 | 2.52 | 1.30e-02 | 7.40e-02 | 8UCHL1, GREM2, RTN1, PLCB1, MAB21L2, LRRN3, GSG1L, SNCG |
389 |
| DURANTE_ADULT_OLFACTORY_NEUROEPITHELIUM_OLFACTORY_ENSHEATHING_GLIA | 1.08e-03 | 9.79 | 2.52 | 8.03e-02 | 7.23e-01 | 4NPY, NRXN1, NR2F1, SNCG |
115 |
| DESCARTES_FETAL_STOMACH_ENS_NEURONS | 3.11e-03 | 11.23 | 2.19 | 1.39e-01 | 1.00e+00 | 3NOS1, NPY, UCHL1 |
74 |
| DURANTE_ADULT_OLFACTORY_NEUROEPITHELIUM_IMMATURE_NEURONS | 2.15e-03 | 8.05 | 2.08 | 1.03e-01 | 1.00e+00 | 4CALM1, UCHL1, RTN1, NRXN1 |
139 |
| FAN_OVARY_CL16_LYMPHATIC_ENDOTHELIAL_CELL | 2.06e-03 | 6.12 | 1.88 | 1.03e-01 | 1.00e+00 | 5CALM1, NTS, CTHRC1, ADM, RTN4 |
232 |
| DESCARTES_FETAL_INTESTINE_ENS_NEURONS | 3.55e-03 | 6.97 | 1.80 | 1.49e-01 | 1.00e+00 | 4CRABP1, NOS1, CNR1, UCHL1 |
160 |
| DURANTE_ADULT_OLFACTORY_NEUROEPITHELIUM_GLOBOSE_BASAL_CELLS | 8.47e-03 | 15.77 | 1.78 | 2.27e-01 | 1.00e+00 | 2UCHL1, RTN1 |
35 |
| HU_FETAL_RETINA_RGC | 1.44e-03 | 4.67 | 1.76 | 8.69e-02 | 9.63e-01 | 7CRABP1, CNR1, UCHL1, RTN1, LRRN3, SNCG, RTN4 |
443 |
| MENON_FETAL_KIDNEY_6_COLLECTING_DUCT_CELLS | 6.06e-03 | 8.76 | 1.71 | 2.01e-01 | 1.00e+00 | 3SPATS2L, CALM1, RTN4 |
94 |
| DURANTE_ADULT_OLFACTORY_NEUROEPITHELIUM_UNSPECIFIED | 9.43e-03 | 14.87 | 1.69 | 2.34e-01 | 1.00e+00 | 2CALM1, NTS |
37 |
| MANNO_MIDBRAIN_NEUROTYPES_HDA1 | 1.50e-03 | 4.14 | 1.67 | 8.69e-02 | 1.00e+00 | 8CNR1, UCHL1, NNAT, RTN1, NRXN1, PLCB1, NAP1L5, MMD |
584 |
Dowload full table
mSigDB Hallmark Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| HALLMARK_INTERFERON_ALPHA_RESPONSE | 2.11e-06 | 18.72 | 6.36 | 5.27e-05 | 1.05e-04 | 6IFI44, IFIT3, IFI27, EIF2AK2, HERC6, LGALS3BP |
97 |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | 6.91e-08 | 14.37 | 6.05 | 3.45e-06 | 3.45e-06 | 9IFIT1, IFI44, IFIT3, IFI27, STAT1, EIF2AK2, OAS3, HERC6, LGALS3BP |
200 |
| HALLMARK_APOPTOSIS | 1.32e-01 | 3.28 | 0.38 | 1.00e+00 | 1.00e+00 | 2HGF, PMAIP1 |
161 |
| HALLMARK_INFLAMMATORY_RESPONSE | 1.85e-01 | 2.63 | 0.31 | 1.00e+00 | 1.00e+00 | 2EIF2AK2, ADM |
200 |
| HALLMARK_HEDGEHOG_SIGNALING | 1.34e-01 | 7.29 | 0.18 | 1.00e+00 | 1.00e+00 | 1RTN1 |
36 |
| HALLMARK_IL6_JAK_STAT3_SIGNALING | 2.93e-01 | 2.97 | 0.07 | 1.00e+00 | 1.00e+00 | 1STAT1 |
87 |
| HALLMARK_PEROXISOME | 3.39e-01 | 2.48 | 0.06 | 1.00e+00 | 1.00e+00 | 1CRABP1 |
104 |
| HALLMARK_PI3K_AKT_MTOR_SIGNALING | 3.41e-01 | 2.45 | 0.06 | 1.00e+00 | 1.00e+00 | 1PLCB1 |
105 |
| HALLMARK_UNFOLDED_PROTEIN_RESPONSE | 3.62e-01 | 2.28 | 0.06 | 1.00e+00 | 1.00e+00 | 1IFIT1 |
113 |
| HALLMARK_SPERMATOGENESIS | 4.15e-01 | 1.90 | 0.05 | 1.00e+00 | 1.00e+00 | 1NOS1 |
135 |
| HALLMARK_HYPOXIA | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1ADM |
200 |
| HALLMARK_ADIPOGENESIS | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1SNCG |
200 |
| HALLMARK_ESTROGEN_RESPONSE_EARLY | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1PMAIP1 |
200 |
| HALLMARK_MYOGENESIS | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1NOS1 |
200 |
| HALLMARK_COMPLEMENT | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1CALM1 |
200 |
| HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1CTHRC1 |
200 |
| HALLMARK_ALLOGRAFT_REJECTION | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1STAT1 |
200 |
| HALLMARK_KRAS_SIGNALING_UP | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1MMD |
200 |
| HALLMARK_KRAS_SIGNALING_DN | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1NOS1 |
200 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
200 |
Dowload full table
KEGG Pathways:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| KEGG_ALZHEIMERS_DISEASE | 2.74e-02 | 4.89 | 0.96 | 1.00e+00 | 1.00e+00 | 3NOS1, CALM1, PLCB1 |
166 |
| KEGG_CALCIUM_SIGNALING_PATHWAY | 3.26e-02 | 4.56 | 0.90 | 1.00e+00 | 1.00e+00 | 3NOS1, CALM1, PLCB1 |
178 |
| KEGG_LONG_TERM_POTENTIATION | 3.13e-02 | 7.66 | 0.88 | 1.00e+00 | 1.00e+00 | 2CALM1, PLCB1 |
70 |
| KEGG_LONG_TERM_DEPRESSION | 3.13e-02 | 7.66 | 0.88 | 1.00e+00 | 1.00e+00 | 2NOS1, PLCB1 |
70 |
| KEGG_MELANOMA | 3.21e-02 | 7.55 | 0.87 | 1.00e+00 | 1.00e+00 | 2FGF10, HGF |
71 |
| KEGG_PHOSPHATIDYLINOSITOL_SIGNALING_SYSTEM | 3.64e-02 | 7.04 | 0.81 | 1.00e+00 | 1.00e+00 | 2CALM1, PLCB1 |
76 |
| KEGG_GNRH_SIGNALING_PATHWAY | 6.04e-02 | 5.26 | 0.61 | 1.00e+00 | 1.00e+00 | 2CALM1, PLCB1 |
101 |
| KEGG_MELANOGENESIS | 6.04e-02 | 5.26 | 0.61 | 1.00e+00 | 1.00e+00 | 2CALM1, PLCB1 |
101 |
| KEGG_VASCULAR_SMOOTH_MUSCLE_CONTRACTION | 7.56e-02 | 4.61 | 0.54 | 1.00e+00 | 1.00e+00 | 2CALM1, PLCB1 |
115 |
| KEGG_PATHWAYS_IN_CANCER | 1.32e-01 | 2.48 | 0.49 | 1.00e+00 | 1.00e+00 | 3STAT1, FGF10, HGF |
325 |
| KEGG_CHEMOKINE_SIGNALING_PATHWAY | 1.70e-01 | 2.79 | 0.33 | 1.00e+00 | 1.00e+00 | 2STAT1, PLCB1 |
189 |
| KEGG_GLYCOSAMINOGLYCAN_BIOSYNTHESIS_CHONDROITIN_SULFATE | 8.40e-02 | 12.14 | 0.29 | 1.00e+00 | 1.00e+00 | 1CSGALNACT1 |
22 |
| KEGG_AMYOTROPHIC_LATERAL_SCLEROSIS_ALS | 1.90e-01 | 4.91 | 0.12 | 1.00e+00 | 1.00e+00 | 1NOS1 |
53 |
| KEGG_ARGININE_AND_PROLINE_METABOLISM | 1.94e-01 | 4.81 | 0.12 | 1.00e+00 | 1.00e+00 | 1NOS1 |
54 |
| KEGG_INOSITOL_PHOSPHATE_METABOLISM | 1.94e-01 | 4.81 | 0.12 | 1.00e+00 | 1.00e+00 | 1PLCB1 |
54 |
| KEGG_GLIOMA | 2.28e-01 | 3.99 | 0.10 | 1.00e+00 | 1.00e+00 | 1CALM1 |
65 |
| KEGG_ADIPOCYTOKINE_SIGNALING_PATHWAY | 2.34e-01 | 3.86 | 0.09 | 1.00e+00 | 1.00e+00 | 1NPY |
67 |
| KEGG_P53_SIGNALING_PATHWAY | 2.37e-01 | 3.81 | 0.09 | 1.00e+00 | 1.00e+00 | 1PMAIP1 |
68 |
| KEGG_RENAL_CELL_CARCINOMA | 2.43e-01 | 3.70 | 0.09 | 1.00e+00 | 1.00e+00 | 1HGF |
70 |
| KEGG_PANCREATIC_CANCER | 2.43e-01 | 3.70 | 0.09 | 1.00e+00 | 1.00e+00 | 1STAT1 |
70 |
Dowload full table
CHR Positional Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| chr4q22 | 3.13e-02 | 7.66 | 0.88 | 1.00e+00 | 1.00e+00 | 2NAP1L5, HERC6 |
70 |
| chr10q23 | 3.84e-02 | 4.27 | 0.84 | 1.00e+00 | 1.00e+00 | 3IFIT1, IFIT3, SNCG |
190 |
| chr2p16 | 7.44e-02 | 4.65 | 0.54 | 1.00e+00 | 1.00e+00 | 2NRXN1, RTN4 |
114 |
| chr12q21 | 9.06e-02 | 4.13 | 0.48 | 1.00e+00 | 1.00e+00 | 2NTS, MGAT4C |
128 |
| chr5q31 | 3.08e-01 | 1.83 | 0.21 | 1.00e+00 | 1.00e+00 | 2PITX1, PCDHB6 |
287 |
| chr4p13 | 1.16e-01 | 8.50 | 0.20 | 1.00e+00 | 1.00e+00 | 1UCHL1 |
31 |
| chr5p12 | 1.20e-01 | 8.23 | 0.20 | 1.00e+00 | 1.00e+00 | 1FGF10 |
32 |
| chr12q24 | 6.63e-01 | 1.34 | 0.16 | 1.00e+00 | 1.00e+00 | 2NOS1, OAS3 |
390 |
| chr6q15 | 1.74e-01 | 5.43 | 0.13 | 1.00e+00 | 1.00e+00 | 1CNR1 |
48 |
| chr6q13 | 2.00e-01 | 4.64 | 0.11 | 1.00e+00 | 1.00e+00 | 1EEF1A1 |
56 |
| chr14q32 | 1.00e+00 | 0.96 | 0.11 | 1.00e+00 | 1.00e+00 | 2IFI27, CALM1 |
546 |
| chr1q43 | 2.13e-01 | 4.32 | 0.11 | 1.00e+00 | 1.00e+00 | 1GREM2 |
60 |
| chr5q15 | 2.13e-01 | 4.32 | 0.11 | 1.00e+00 | 1.00e+00 | 1NR2F1 |
60 |
| chr6q24 | 2.49e-01 | 3.59 | 0.09 | 1.00e+00 | 1.00e+00 | 1AIG1 |
72 |
| chr2p24 | 2.55e-01 | 3.49 | 0.09 | 1.00e+00 | 1.00e+00 | 1NBAS |
74 |
| chr2p21 | 2.73e-01 | 3.23 | 0.08 | 1.00e+00 | 1.00e+00 | 1THADA |
80 |
| chrXp21 | 2.78e-01 | 3.15 | 0.08 | 1.00e+00 | 1.00e+00 | 1ARX |
82 |
| chr17q22 | 2.90e-01 | 3.00 | 0.07 | 1.00e+00 | 1.00e+00 | 1MMD |
86 |
| chr7p15 | 3.17e-01 | 2.68 | 0.07 | 1.00e+00 | 1.00e+00 | 1NPY |
96 |
| chr2p22 | 3.23e-01 | 2.63 | 0.06 | 1.00e+00 | 1.00e+00 | 1EIF2AK2 |
98 |
Dowload full table
Transcription Factor Targets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| MSX1_01 | 5.98e-05 | 10.09 | 3.46 | 6.77e-02 | 6.77e-02 | 6CALM1, NTS, NR2F1, ARX, MAB21L2, AIG1 |
175 |
| RNTCANNRNNYNATTW_UNKNOWN | 2.06e-03 | 13.07 | 2.53 | 5.14e-01 | 1.00e+00 | 3FGF10, NRXN1, MGAT4C |
64 |
| MEF2_04 | 4.74e-03 | 21.66 | 2.42 | 5.37e-01 | 1.00e+00 | 2NR2F1, CTHRC1 |
26 |
| GRE_C | 1.64e-03 | 8.69 | 2.24 | 5.14e-01 | 1.00e+00 | 4NNAT, NR2F1, ARX, LRRN3 |
129 |
| PER1_TARGET_GENES | 7.55e-03 | 16.78 | 1.89 | 6.43e-01 | 1.00e+00 | 2NTS, EEF1A1 |
33 |
| TTCNRGNNNNTTC_HSF_Q6 | 3.17e-03 | 7.20 | 1.86 | 5.14e-01 | 1.00e+00 | 4FGF10, NNAT, RTN1, NR2F1 |
155 |
| CP2_02 | 2.97e-03 | 5.60 | 1.72 | 5.14e-01 | 1.00e+00 | 5NNAT, RTN1, NR2F1, ARX, CUTA |
253 |
| OCT1_07 | 4.22e-03 | 6.63 | 1.71 | 5.31e-01 | 1.00e+00 | 4NOS1, PLCB1, NR2F1, MGAT4C |
168 |
| TGANTCA_AP1_C | 6.46e-04 | 3.51 | 1.66 | 3.66e-01 | 7.31e-01 | 12STAT1, FGF10, RTN1, NRXN1, ARX, EEF1A1, AIG1, LRRN3, CTHRC1, SNCG, ADM, RTN4 |
1139 |
| HNF4ALPHA_Q6 | 3.96e-03 | 5.22 | 1.60 | 5.31e-01 | 1.00e+00 | 5FGF10, NR2F1, MAB21L2, CUTA, RTN4 |
271 |
| STTTCRNTTT_IRF_Q6 | 6.70e-03 | 5.78 | 1.50 | 6.33e-01 | 1.00e+00 | 4IFI44, IFIT3, HGF, LGALS3BP |
192 |
| TGGAAA_NFAT_Q4_01 | 2.42e-03 | 2.79 | 1.42 | 5.14e-01 | 1.00e+00 | 15NOS1, FGF10, CALM1, CNR1, UCHL1, HGF, NNAT, PLCB1, MAB21L2, AIG1, LRRN3, CTHRC1, TMEM108, NAP1L5, ADM |
1934 |
| RP58_01 | 9.39e-03 | 5.22 | 1.35 | 6.49e-01 | 1.00e+00 | 4FGF10, CALM1, HGF, ARX |
212 |
| TGCCAAR_NF1_Q6 | 5.41e-03 | 3.31 | 1.34 | 5.57e-01 | 1.00e+00 | 8CALM1, HGF, NNAT, PLCB1, NR2F1, MGAT4C, MAB21L2, PITX1 |
727 |
| YGCANTGCR_UNKNOWN | 1.37e-02 | 6.43 | 1.26 | 6.49e-01 | 1.00e+00 | 3HGF, NNAT, NRXN1 |
127 |
| CTGCAGY_UNKNOWN | 7.94e-03 | 3.09 | 1.25 | 6.43e-01 | 1.00e+00 | 8CALM1, HGF, NNAT, NRXN1, NR2F1, MGAT4C, TMEM108, RTN4 |
779 |
| NKX3A_01 | 1.38e-02 | 4.65 | 1.20 | 6.49e-01 | 1.00e+00 | 4NNAT, NR2F1, ARX, NAP1L5 |
238 |
| YATGNWAAT_OCT_C | 1.34e-02 | 3.84 | 1.18 | 6.49e-01 | 1.00e+00 | 5NOS1, NRXN1, MAB21L2, NAP1L5, ADM |
367 |
| SMAD4_Q6 | 1.50e-02 | 4.53 | 1.17 | 6.49e-01 | 1.00e+00 | 4CALM1, NR2F1, MGAT4C, EEF1A1 |
244 |
| IRF_Q6 | 1.50e-02 | 4.53 | 1.17 | 6.49e-01 | 1.00e+00 | 4IFI44, HMCN1, TMEM108, RTN4 |
244 |
Dowload full table
GO Biological Processes:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GOBP_RETROGRADE_TRANS_SYNAPTIC_SIGNALING | 4.91e-06 | 132.55 | 20.77 | 3.67e-02 | 3.67e-02 | 3NOS1, CNR1, PLCB1 |
9 |
| GOBP_RETROGRADE_TRANS_SYNAPTIC_SIGNALING_BY_LIPID | 1.54e-04 | 172.68 | 14.09 | 2.31e-01 | 1.00e+00 | 2CNR1, PLCB1 |
5 |
| GOBP_REGULATION_BY_VIRUS_OF_VIRAL_PROTEIN_LEVELS_IN_HOST_CELL | 3.22e-04 | 103.49 | 9.64 | 2.61e-01 | 1.00e+00 | 2IFIT1, STAT1 |
7 |
| GOBP_MESENCHYMAL_EPITHELIAL_CELL_SIGNALING | 3.22e-04 | 103.49 | 9.64 | 2.61e-01 | 1.00e+00 | 2FGF10, HGF |
7 |
| GOBP_REGULATION_OF_BRANCHING_INVOLVED_IN_SALIVARY_GLAND_MORPHOGENESIS | 3.22e-04 | 103.49 | 9.64 | 2.61e-01 | 1.00e+00 | 2FGF10, HGF |
7 |
| GOBP_TRANS_SYNAPTIC_SIGNALING_BY_LIPID | 3.22e-04 | 103.49 | 9.64 | 2.61e-01 | 1.00e+00 | 2CNR1, PLCB1 |
7 |
| GOBP_EMBRYONIC_BODY_MORPHOGENESIS | 8.35e-04 | 57.69 | 5.91 | 4.16e-01 | 1.00e+00 | 2GREM2, MAB21L2 |
11 |
| GOBP_RESPONSE_TO_TYPE_I_INTERFERON | 4.68e-05 | 14.46 | 4.38 | 1.17e-01 | 3.50e-01 | 5IFIT1, IFIT3, IFI27, STAT1, OAS3 |
101 |
| GOBP_NEGATIVE_REGULATION_OF_CALCIUM_ION_TRANSPORT_INTO_CYTOSOL | 2.03e-03 | 34.68 | 3.75 | 5.84e-01 | 1.00e+00 | 2NOS1, CALM1 |
17 |
| GOBP_REGULATION_OF_MORPHOGENESIS_OF_A_BRANCHING_STRUCTURE | 8.97e-04 | 17.71 | 3.40 | 4.20e-01 | 1.00e+00 | 3FGF10, HGF, RTN4 |
48 |
| GOBP_AXONAL_FASCICULATION | 2.54e-03 | 30.58 | 3.34 | 7.02e-01 | 1.00e+00 | 2CNR1, RTN4 |
19 |
| GOBP_NEGATIVE_REGULATION_OF_VIRAL_PROCESS | 4.13e-04 | 12.78 | 3.27 | 2.61e-01 | 1.00e+00 | 4IFIT1, STAT1, EIF2AK2, OAS3 |
89 |
| GOBP_SEROTONIN_TRANSPORT | 2.81e-03 | 28.89 | 3.17 | 7.02e-01 | 1.00e+00 | 2NOS1, CNR1 |
20 |
| GOBP_BRANCHING_INVOLVED_IN_SALIVARY_GLAND_MORPHOGENESIS | 2.81e-03 | 28.89 | 3.17 | 7.02e-01 | 1.00e+00 | 2FGF10, HGF |
20 |
| GOBP_DEFENSE_RESPONSE_TO_VIRUS | 6.21e-05 | 8.04 | 3.02 | 1.17e-01 | 4.64e-01 | 7IFIT1, IFIT3, IFI27, STAT1, EIF2AK2, PMAIP1, OAS3 |
260 |
| GOBP_NEGATIVE_REGULATION_OF_VIRAL_GENOME_REPLICATION | 1.26e-03 | 15.63 | 3.02 | 5.05e-01 | 1.00e+00 | 3IFIT1, EIF2AK2, OAS3 |
54 |
| GOBP_RESPONSE_TO_INTERFERON_ALPHA | 3.10e-03 | 27.37 | 3.01 | 7.20e-01 | 1.00e+00 | 2IFIT3, EIF2AK2 |
21 |
| GOBP_REGULATION_OF_NEUROTRANSMITTER_TRANSPORT | 6.17e-04 | 11.44 | 2.93 | 3.55e-01 | 1.00e+00 | 4NOS1, CALM1, CNR1, SNCG |
99 |
| GOBP_PALLIUM_DEVELOPMENT | 3.55e-04 | 9.20 | 2.81 | 2.61e-01 | 1.00e+00 | 5NPY, PLCB1, ARX, TMEM108, RTN4 |
156 |
| GOBP_RESPONSE_TO_VIRUS | 6.28e-05 | 6.80 | 2.74 | 1.17e-01 | 4.70e-01 | 8IFIT1, IFI44, IFIT3, IFI27, STAT1, EIF2AK2, PMAIP1, OAS3 |
358 |
Dowload full table
Immunological Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GSE10325_CD4_TCELL_VS_LUPUS_CD4_TCELL_DN | 1.08e-11 | 21.10 | 9.88 | 5.29e-08 | 5.29e-08 | 12IFIT1, IFI44, IFIT3, IFI27, STAT1, SPATS2L, EIF2AK2, PMAIP1, OAS3, HERC6, LGALS3BP, MMD |
199 |
| GSE6269_HEALTHY_VS_FLU_INF_PBMC_DN | 6.02e-10 | 20.40 | 8.94 | 5.86e-07 | 2.93e-06 | 10IFIT1, IFI44, IFIT3, IFI27, SPATS2L, EIF2AK2, OAS3, HERC6, LGALS3BP, ADM |
163 |
| GSE42724_NAIVE_BCELL_VS_PLASMABLAST_UP | 2.01e-10 | 18.95 | 8.62 | 2.87e-07 | 9.78e-07 | 11IFIT1, IFI44, IFIT3, IFI27, STAT1, SPATS2L, EIF2AK2, RTN1, OAS3, HERC6, LGALS3BP |
197 |
| GSE10325_MYELOID_VS_LUPUS_MYELOID_DN | 2.35e-10 | 18.65 | 8.48 | 2.87e-07 | 1.15e-06 | 11IFIT1, IFI44, IFIT3, IFI27, STAT1, SPATS2L, EIF2AK2, CALM1, HERC6, LGALS3BP, ADM |
200 |
| GSE42021_TREG_VS_TCONV_PLN_UP | 2.35e-10 | 18.65 | 8.48 | 2.87e-07 | 1.15e-06 | 11IFIT1, IFI44, IFIT3, IFI27, STAT1, SPATS2L, EIF2AK2, PMAIP1, OAS3, HERC6, LGALS3BP |
200 |
| GSE21546_UNSTIM_VS_ANTI_CD3_STIM_ELK1_KO_DP_THYMOCYTES_UP | 2.63e-09 | 17.36 | 7.62 | 1.31e-06 | 1.28e-05 | 10IFIT1, IFI44, IFIT3, IFI27, STAT1, EIF2AK2, RTN1, OAS3, HERC6, LGALS3BP |
190 |
| GSE21546_WT_VS_SAP1A_KO_DP_THYMOCYTES_UP | 3.54e-09 | 16.80 | 7.38 | 1.31e-06 | 1.72e-05 | 10IFIT1, IFI44, IFIT3, IFI27, STAT1, SPATS2L, EIF2AK2, OAS3, HERC6, LGALS3BP |
196 |
| GSE42021_TREG_PLN_VS_CD24LO_TREG_THYMUS_DN | 3.72e-09 | 16.71 | 7.34 | 1.31e-06 | 1.81e-05 | 10IFIT1, IFI44, IFIT3, IFI27, STAT1, SPATS2L, EIF2AK2, OAS3, HERC6, LGALS3BP |
197 |
| GSE13485_DAY1_VS_DAY7_YF17D_VACCINE_PBMC_DN | 3.90e-09 | 16.62 | 7.30 | 1.31e-06 | 1.90e-05 | 10IFIT1, IFI44, IFIT3, IFI27, STAT1, SPATS2L, EIF2AK2, OAS3, HERC6, LGALS3BP |
198 |
| GSE13485_DAY3_VS_DAY7_YF17D_VACCINE_PBMC_DN | 3.90e-09 | 16.62 | 7.30 | 1.31e-06 | 1.90e-05 | 10IFIT1, IFI44, IFIT3, IFI27, STAT1, SPATS2L, EIF2AK2, OAS3, HERC6, LGALS3BP |
198 |
| GSE13485_PRE_VS_POST_YF17D_VACCINATION_PBMC_DN | 3.90e-09 | 16.62 | 7.30 | 1.31e-06 | 1.90e-05 | 10IFIT1, IFI44, IFIT3, IFI27, STAT1, SPATS2L, EIF2AK2, OAS3, HERC6, LGALS3BP |
198 |
| GSE13484_UNSTIM_VS_YF17D_VACCINE_STIM_PBMC_DN | 4.29e-09 | 16.45 | 7.22 | 1.31e-06 | 2.09e-05 | 10IFIT1, IFI44, IFIT3, STAT1, SPATS2L, EIF2AK2, CALM1, OAS3, HERC6, LGALS3BP |
200 |
| GSE13485_CTRL_VS_DAY7_YF17D_VACCINE_PBMC_DN | 4.29e-09 | 16.45 | 7.22 | 1.31e-06 | 2.09e-05 | 10IFIT1, IFI44, IFIT3, IFI27, STAT1, SPATS2L, EIF2AK2, OAS3, HERC6, LGALS3BP |
200 |
| GSE22886_CTRL_VS_LPS_24H_DC_DN | 4.29e-09 | 16.45 | 7.22 | 1.31e-06 | 2.09e-05 | 10IFIT1, IFI44, IFIT3, IFI27, STAT1, EIF2AK2, PMAIP1, OAS3, HERC6, LGALS3BP |
200 |
| GSE26030_TH1_VS_TH17_DAY5_POST_POLARIZATION_UP | 4.29e-09 | 16.45 | 7.22 | 1.31e-06 | 2.09e-05 | 10IFIT1, IFI44, IFIT3, STAT1, EIF2AK2, PKIB, PITX1, OAS3, HERC6, LGALS3BP |
200 |
| GSE42021_TREG_PLN_VS_TREG_PRECURSORS_THYMUS_DN | 4.29e-09 | 16.45 | 7.22 | 1.31e-06 | 2.09e-05 | 10IFIT1, IFI44, IFIT3, IFI27, STAT1, SPATS2L, EIF2AK2, OAS3, HERC6, LGALS3BP |
200 |
| GSE6269_FLU_VS_STREP_PNEUMO_INF_PBMC_UP | 1.81e-08 | 16.93 | 7.12 | 5.20e-06 | 8.83e-05 | 9IFIT1, IFI44, IFIT3, STAT1, SPATS2L, EIF2AK2, OAS3, HERC6, LGALS3BP |
171 |
| GSE21546_UNSTIM_VS_ANTI_CD3_STIM_SAP1A_KO_AND_ELK1_KO_DP_THYMOCYTES_UP | 4.27e-08 | 15.24 | 6.41 | 1.16e-05 | 2.08e-04 | 9IFIT1, IFIT3, IFI27, STAT1, EIF2AK2, MGAT4C, OAS3, HERC6, LGALS3BP |
189 |
| GSE37533_PPARG2_FOXP3_VS_FOXP3_TRANSDUCED_CD4_TCELL_DN | 6.34e-08 | 14.52 | 6.11 | 1.35e-05 | 3.09e-04 | 9IFIT1, IFI44, IFIT3, STAT1, SPATS2L, EIF2AK2, PMAIP1, OAS3, HERC6 |
198 |
| GSE13485_CTRL_VS_DAY3_YF17D_VACCINE_PBMC_DN | 6.62e-08 | 14.44 | 6.08 | 1.35e-05 | 3.22e-04 | 9IFIT1, IFI44, IFIT3, IFI27, STAT1, SPATS2L, EIF2AK2, OAS3, HERC6 |
199 |
Top Ranked Transcription Factors for this Gene Expression Program:
| Gene Symbol | TF Rank | DNA Binding Domain | Motif Status | IUPAC PWM | GTEx | DepMap | Decartes |
|---|---|---|---|---|---|---|---|
| STAT1 | 6 | Yes | Known motif | Monomer or homomultimer | In vivo/Misc source | None | Structure PDB:1BF5 is Tyrosine phosphorylated |
| EIF2AK2 | 10 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| NR2F1 | 26 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| ARX | 28 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| PITX1 | 37 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| CDC5L | 53 | Yes | Known motif | Monomer or homomultimer | In vivo/Misc source | Only known motifs are from Transfac or HocoMoco - origin is uncertain | Compelling evidence for DNA binding (PMID: 11082045). |
| PLSCR1 | 58 | Yes | Likely to be sequence specific TF | Monomer or homomultimer | No motif | None | PLSCR1 has been demonstrated to bind a GTAACCATGTGGA sequence in the IP3R1 promoter (PMID:16091359), mediated by a specific portion of the protein. |
| TWIST1 | 68 | Yes | Known motif | Monomer or homomultimer | In vivo/Misc source | Only known motifs are from Transfac or HocoMoco - origin is uncertain | Can form both homodimers and heterodimers with TCF3 (PMID: 16502419). |
| TBX3 | 75 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| HDAC9 | 80 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Histone deacetylase; likely to be a transcriptional cofactor |
| DDX58 | 84 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| YBX1 | 87 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | Might also bind RNA |
| HMBOX1 | 102 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| PRRX2 | 105 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| AFF3 | 106 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | (PMID: 8555498) only says that In vitro-translated LAF-4 was able to bind strongly to double-stranded DNA cellulose - thats more consistent with nonspecific DNA binding. The key issue is whether the Transcription factor AF4/FMR2 (IPR007797) domain really is sequence specific. Most of the work has been done on the one Drosophila ortholog, lilliuputian. I see no evidence of DNA binding. Recent work on this family in human indicate that it functions in coactivation (PMID: 27899651; PMID: 261712 |
| SCOC | 110 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Not a proper bZIP; contains only a small leucine zipper region but lacks the DNA-interfacing basic part. |
| BNC1 | 123 | Yes | Inferred motif | Monomer or homomultimer | In vivo/Misc source | None | None |
| MBNL2 | 138 | Yes | Likely to be sequence specific TF | Monomer or homomultimer | High-throughput in vitro | None | Has methylated and unmethylated HT-SELEX motifs, despite this family largely binding RNA exclusively (PMID: 28473536 ) |
| EDF1 | 152 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Known co-factor that binds DNA indirectly through interactions with TFs (PMID:12040021) |
| TEAD1 | 177 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
QQ Plot showing correlations with other GEPs in this dataset, calculated by Spearman correlation:
Interactive QQ-plot of gene loadings:
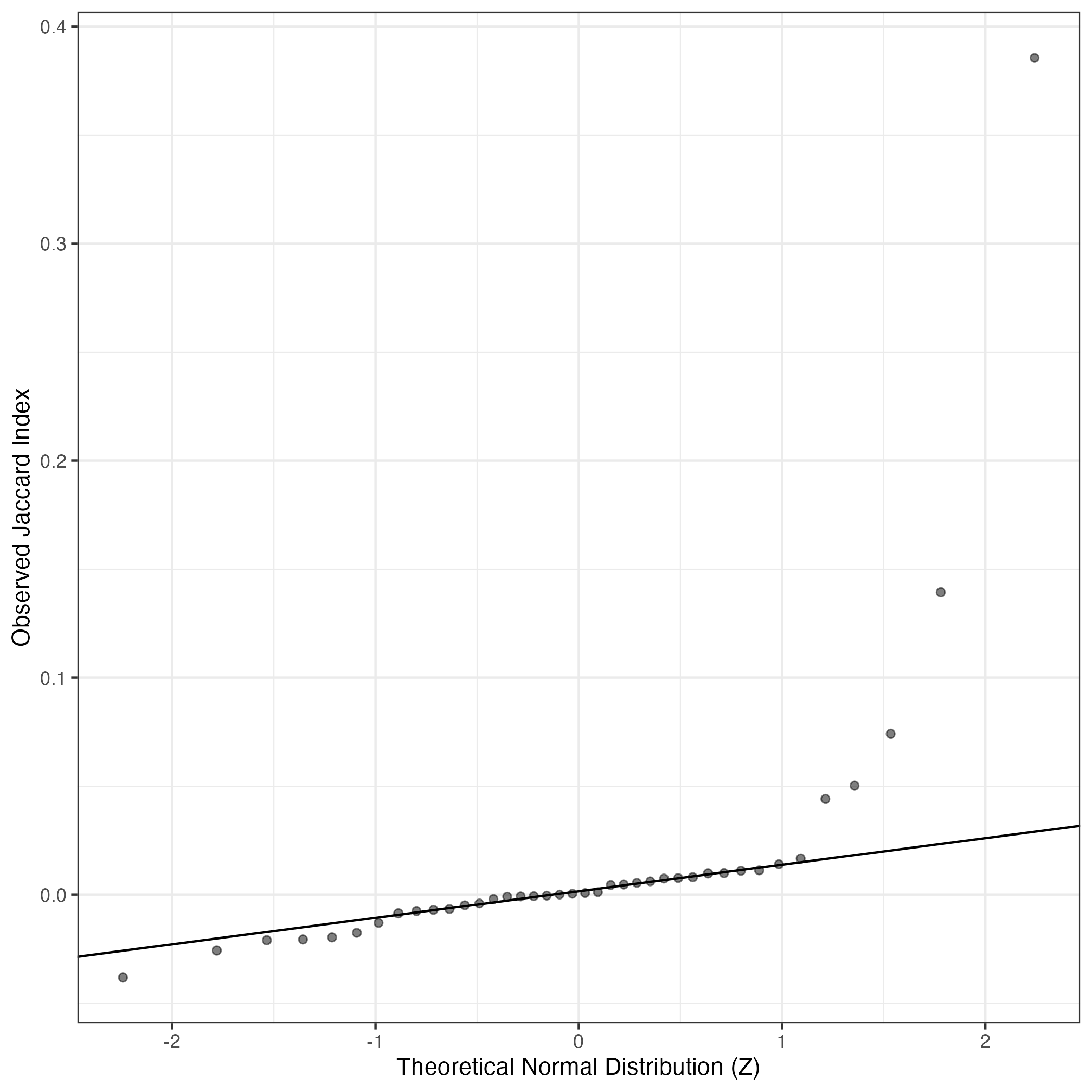
A similar QQ-plot as above, but only for instances where the H value is e.g. > 25, i.e. we are confident that the expression program is active above noise. Agreemenet between these binary vectors is tested using the Jaccard Index, with the P-values calculated by an exact test:
Interactive QQ-plot:
Singler cell type annotations for the top 50 cells on this program.
| Cell ID | Singler label | Singler Delta | Activity Score | Top Singler Raw Scores |
|---|---|---|---|---|
| T200_AATAGAGTCGCAATTG-1 | Neurons:adrenal_medulla_cell_line | 0.13 | 724.10 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.29, Astrocyte:Embryonic_stem_cell-derived: 0.27, Neuroepithelial_cell:ESC-derived: 0.27, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.24, iPS_cells:PDB_1lox-21Puro-20: 0.23, Neurons:ES_cell-derived_neural_precursor: 0.23, iPS_cells:PDB_1lox-17Puro-10: 0.23, iPS_cells:PDB_1lox-17Puro-5: 0.23, iPS_cells:PDB_2lox-22: 0.23, iPS_cells:adipose_stem_cell-derived:lentiviral: 0.23 |
| T200_GTGTTCCGTAGGTGCA-1 | Neurons:adrenal_medulla_cell_line | 0.13 | 719.76 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Neuroepithelial_cell:ESC-derived: 0.27, Astrocyte:Embryonic_stem_cell-derived: 0.27, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.26, iPS_cells:PDB_1lox-17Puro-5: 0.25, iPS_cells:PDB_1lox-21Puro-20: 0.25, iPS_cells:adipose_stem_cell-derived:lentiviral: 0.25, iPS_cells:PDB_1lox-17Puro-10: 0.25, iPS_cells:skin_fibroblast-derived: 0.25, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.25 |
| T200_TTGTTTGAGTTACGTC-1 | Neurons:adrenal_medulla_cell_line | 0.13 | 713.97 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Neuroepithelial_cell:ESC-derived: 0.27, Astrocyte:Embryonic_stem_cell-derived: 0.27, iPS_cells:PDB_1lox-17Puro-5: 0.26, Embryonic_stem_cells: 0.26, iPS_cells:PDB_2lox-22: 0.25, iPS_cells:PDB_1lox-21Puro-26: 0.25, iPS_cells:PDB_1lox-21Puro-20: 0.25, iPS_cells:PDB_2lox-17: 0.25, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.25 |
| T200_AGGCTGCTCTGGCCAG-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 697.29 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.32, Neuroepithelial_cell:ESC-derived: 0.29, Astrocyte:Embryonic_stem_cell-derived: 0.28, iPS_cells:PDB_1lox-17Puro-10: 0.27, iPS_cells:PDB_1lox-17Puro-5: 0.26, Neurons:ES_cell-derived_neural_precursor: 0.26, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.26, iPS_cells:PDB_1lox-21Puro-20: 0.26, iPS_cells:PDB_1lox-21Puro-26: 0.26, iPS_cells:PDB_2lox-22: 0.26 |
| T200_GGCTGTGTCGATACGT-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 650.91 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.32, Astrocyte:Embryonic_stem_cell-derived: 0.28, Neuroepithelial_cell:ESC-derived: 0.27, Neurons:Schwann_cell: 0.25, iPS_cells:PDB_1lox-17Puro-5: 0.25, iPS_cells:PDB_1lox-17Puro-10: 0.25, iPS_cells:PDB_1lox-21Puro-20: 0.24, Smooth_muscle_cells:bronchial: 0.24, iPS_cells:PDB_1lox-21Puro-26: 0.24, MSC: 0.24 |
| T200_GGGAAGTCACAAACGG-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 639.52 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.32, Astrocyte:Embryonic_stem_cell-derived: 0.3, Neuroepithelial_cell:ESC-derived: 0.29, Embryonic_stem_cells: 0.27, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.27, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.26, Fibroblasts:breast: 0.26, MSC: 0.26, Smooth_muscle_cells:umbilical_vein: 0.26, iPS_cells:PDB_2lox-5: 0.26 |
| T200_GTGGTTATCATCGCTC-1 | Neurons:adrenal_medulla_cell_line | 0.21 | 619.26 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.39, Neuroepithelial_cell:ESC-derived: 0.36, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.34, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_2lox-22: 0.34, iPS_cells:PDB_2lox-21: 0.33, Embryonic_stem_cells: 0.33, iPS_cells:PDB_2lox-17: 0.33 |
| T200_CGGTCAGTCAACGTGT-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 609.57 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Astrocyte:Embryonic_stem_cell-derived: 0.27, Neuroepithelial_cell:ESC-derived: 0.27, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.24, iPS_cells:adipose_stem_cell-derived:lentiviral: 0.24, iPS_cells:PDB_1lox-21Puro-20: 0.24, iPS_cells:PDB_2lox-17: 0.24, iPS_cells:PDB_2lox-5: 0.24, iPS_cells:PDB_2lox-22: 0.24, iPS_cells:PDB_2lox-21: 0.24 |
| T200_ATTACCTTCGAAGCCC-1 | Neurons:adrenal_medulla_cell_line | 0.20 | 608.30 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.31, Neuroepithelial_cell:ESC-derived: 0.3, Neurons:ES_cell-derived_neural_precursor: 0.27, Embryonic_stem_cells: 0.27, iPS_cells:PDB_1lox-21Puro-20: 0.27, iPS_cells:PDB_1lox-17Puro-5: 0.27, iPS_cells:PDB_1lox-17Puro-10: 0.27, iPS_cells:PDB_1lox-21Puro-26: 0.27, iPS_cells:PDB_2lox-22: 0.26 |
| T200_TCCGAAACAGCTACCG-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 606.56 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.34, Astrocyte:Embryonic_stem_cell-derived: 0.29, Neuroepithelial_cell:ESC-derived: 0.29, iPS_cells:PDB_1lox-21Puro-20: 0.26, iPS_cells:PDB_1lox-17Puro-5: 0.26, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.26, iPS_cells:PDB_1lox-17Puro-10: 0.26, iPS_cells:PDB_2lox-22: 0.26, iPS_cells:PDB_1lox-21Puro-26: 0.26, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.26 |
| T200_CTGTACCGTATAGCTC-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 549.75 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.34, Neuroepithelial_cell:ESC-derived: 0.3, Astrocyte:Embryonic_stem_cell-derived: 0.3, Neurons:ES_cell-derived_neural_precursor: 0.27, Embryonic_stem_cells: 0.27, iPS_cells:PDB_1lox-17Puro-5: 0.27, iPS_cells:PDB_1lox-17Puro-10: 0.27, iPS_cells:PDB_1lox-21Puro-20: 0.26, iPS_cells:PDB_1lox-21Puro-26: 0.26, iPS_cells:PDB_2lox-22: 0.26 |
| T200_AATGGAATCTTCTGTA-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 544.83 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.34, Astrocyte:Embryonic_stem_cell-derived: 0.31, Neuroepithelial_cell:ESC-derived: 0.3, iPS_cells:PDB_1lox-21Puro-20: 0.26, iPS_cells:PDB_1lox-17Puro-10: 0.26, iPS_cells:PDB_1lox-17Puro-5: 0.26, iPS_cells:PDB_1lox-21Puro-26: 0.26, Neurons:ES_cell-derived_neural_precursor: 0.26, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.26, Embryonic_stem_cells: 0.26 |
| T200_CCCATTGTCGAGAGAC-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 543.08 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.32, Neuroepithelial_cell:ESC-derived: 0.29, Astrocyte:Embryonic_stem_cell-derived: 0.28, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.27, iPS_cells:adipose_stem_cell-derived:lentiviral: 0.26, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.26, iPS_cells:PDB_2lox-5: 0.26, iPS_cells:PDB_2lox-22: 0.26, iPS_cells:iPS:minicircle-derived: 0.25, iPS_cells:skin_fibroblast-derived: 0.25 |
| T200_GCGGAAAAGAAACTCA-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 538.11 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Neuroepithelial_cell:ESC-derived: 0.26, Astrocyte:Embryonic_stem_cell-derived: 0.26, iPS_cells:PDB_1lox-21Puro-20: 0.23, iPS_cells:PDB_1lox-17Puro-5: 0.23, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.23, Neurons:ES_cell-derived_neural_precursor: 0.23, iPS_cells:PDB_2lox-22: 0.23, iPS_cells:PDB_1lox-17Puro-10: 0.22, iPS_cells:PDB_1lox-21Puro-26: 0.22 |
| T200_AGCTCAATCGCTCTAC-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 500.66 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.36, Astrocyte:Embryonic_stem_cell-derived: 0.31, Neuroepithelial_cell:ESC-derived: 0.3, iPS_cells:PDB_1lox-21Puro-20: 0.28, iPS_cells:PDB_1lox-17Puro-5: 0.28, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.28, iPS_cells:PDB_1lox-21Puro-26: 0.28, iPS_cells:PDB_1lox-17Puro-10: 0.28, iPS_cells:PDB_2lox-22: 0.28, iPS_cells:PDB_2lox-21: 0.27 |
| T200_GACCAATCATCAGCGC-1 | Neurons:adrenal_medulla_cell_line | 0.09 | 494.15 | Raw ScoresTissue_stem_cells:BM_MSC:BMP2: 0.27, Fibroblasts:breast: 0.27, Tissue_stem_cells:BM_MSC:TGFb3: 0.27, Neurons:adrenal_medulla_cell_line: 0.27, Astrocyte:Embryonic_stem_cell-derived: 0.27, Smooth_muscle_cells:bronchial: 0.27, Neuroepithelial_cell:ESC-derived: 0.27, iPS_cells:adipose_stem_cells: 0.27, Smooth_muscle_cells:bronchial:vit_D: 0.26, Chondrocytes:MSC-derived: 0.26 |
| T200_CACTGAAGTTCCCACT-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 487.26 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.34, Astrocyte:Embryonic_stem_cell-derived: 0.3, Neuroepithelial_cell:ESC-derived: 0.29, iPS_cells:PDB_1lox-21Puro-20: 0.26, Neurons:ES_cell-derived_neural_precursor: 0.26, iPS_cells:PDB_1lox-17Puro-5: 0.26, iPS_cells:PDB_1lox-17Puro-10: 0.26, iPS_cells:PDB_1lox-21Puro-26: 0.26, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.26, iPS_cells:PDB_2lox-22: 0.26 |
| T200_TCATGCCTCACTTTGT-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 475.36 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Neuroepithelial_cell:ESC-derived: 0.28, Astrocyte:Embryonic_stem_cell-derived: 0.28, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.26, iPS_cells:PDB_1lox-17Puro-10: 0.26, iPS_cells:PDB_1lox-17Puro-5: 0.26, iPS_cells:skin_fibroblast-derived: 0.25, iPS_cells:PDB_1lox-21Puro-20: 0.25, Embryonic_stem_cells: 0.25, iPS_cells:PDB_2lox-5: 0.25 |
| T200_TAACCAGTCTCCTACG-1 | Neurons:adrenal_medulla_cell_line | 0.18 | 474.26 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.32, Neuroepithelial_cell:ESC-derived: 0.28, Astrocyte:Embryonic_stem_cell-derived: 0.28, Neurons:ES_cell-derived_neural_precursor: 0.25, iPS_cells:PDB_2lox-5: 0.24, Embryonic_stem_cells: 0.24, iPS_cells:PDB_1lox-21Puro-26: 0.24, iPS_cells:PDB_1lox-21Puro-20: 0.24, iPS_cells:PDB_1lox-17Puro-10: 0.24, iPS_cells:PDB_1lox-17Puro-5: 0.24 |
| T200_GCCAGTGGTGGTCCCA-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 464.77 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Astrocyte:Embryonic_stem_cell-derived: 0.27, Neuroepithelial_cell:ESC-derived: 0.26, iPS_cells:PDB_1lox-17Puro-5: 0.24, iPS_cells:PDB_1lox-17Puro-10: 0.24, iPS_cells:PDB_1lox-21Puro-20: 0.24, Neurons:ES_cell-derived_neural_precursor: 0.23, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.23, iPS_cells:PDB_1lox-21Puro-26: 0.23, Embryonic_stem_cells: 0.23 |
| T200_GCTCAAATCATGAGAA-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 463.63 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.34, Neuroepithelial_cell:ESC-derived: 0.3, Astrocyte:Embryonic_stem_cell-derived: 0.29, Neurons:ES_cell-derived_neural_precursor: 0.26, iPS_cells:PDB_1lox-21Puro-20: 0.26, iPS_cells:PDB_1lox-17Puro-10: 0.26, iPS_cells:PDB_1lox-21Puro-26: 0.26, iPS_cells:PDB_1lox-17Puro-5: 0.26, iPS_cells:PDB_2lox-22: 0.25, Embryonic_stem_cells: 0.25 |
| T200_CCATAAGGTACTAACC-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 460.67 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Neuroepithelial_cell:ESC-derived: 0.29, Astrocyte:Embryonic_stem_cell-derived: 0.28, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.27, iPS_cells:PDB_1lox-17Puro-5: 0.26, iPS_cells:PDB_1lox-17Puro-10: 0.26, iPS_cells:PDB_1lox-21Puro-20: 0.26, iPS_cells:PDB_1lox-21Puro-26: 0.26, iPS_cells:PDB_2lox-22: 0.26, iPS_cells:PDB_2lox-21: 0.26 |
| T200_GTTACCCAGCATTGAA-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 457.73 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.35, Neuroepithelial_cell:ESC-derived: 0.32, Astrocyte:Embryonic_stem_cell-derived: 0.31, iPS_cells:PDB_1lox-21Puro-20: 0.28, Neurons:ES_cell-derived_neural_precursor: 0.28, iPS_cells:PDB_1lox-17Puro-10: 0.28, iPS_cells:PDB_1lox-17Puro-5: 0.28, iPS_cells:PDB_1lox-21Puro-26: 0.27, Embryonic_stem_cells: 0.27, iPS_cells:PDB_2lox-22: 0.27 |
| T200_TCTATACCAGGTCTCG-1 | Neurons:adrenal_medulla_cell_line | 0.13 | 454.99 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.29, Astrocyte:Embryonic_stem_cell-derived: 0.27, Neuroepithelial_cell:ESC-derived: 0.26, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.24, iPS_cells:PDB_1lox-21Puro-20: 0.24, iPS_cells:PDB_1lox-21Puro-26: 0.24, iPS_cells:PDB_1lox-17Puro-5: 0.24, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.24, iPS_cells:PDB_2lox-5: 0.24, iPS_cells:PDB_2lox-21: 0.24 |
| T200_TACTGCCAGGGAGGAC-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 445.68 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.31, Astrocyte:Embryonic_stem_cell-derived: 0.28, Neuroepithelial_cell:ESC-derived: 0.27, Neurons:ES_cell-derived_neural_precursor: 0.24, Embryonic_stem_cells: 0.24, iPS_cells:PDB_1lox-21Puro-20: 0.23, iPS_cells:PDB_1lox-17Puro-5: 0.23, iPS_cells:PDB_1lox-17Puro-10: 0.23, iPS_cells:PDB_1lox-21Puro-26: 0.23, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.23 |
| T200_CCGATGGCAGTTTCAG-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 437.14 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.31, Neuroepithelial_cell:ESC-derived: 0.27, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.26, iPS_cells:adipose_stem_cell-derived:lentiviral: 0.26, Astrocyte:Embryonic_stem_cell-derived: 0.26, iPS_cells:skin_fibroblast-derived: 0.26, iPS_cells:PDB_1lox-17Puro-10: 0.26, iPS_cells:iPS:minicircle-derived: 0.26, Embryonic_stem_cells: 0.26, iPS_cells:PDB_1lox-21Puro-20: 0.25 |
| T200_GATCACAGTGACTGTT-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 434.81 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Astrocyte:Embryonic_stem_cell-derived: 0.27, Neuroepithelial_cell:ESC-derived: 0.27, iPS_cells:PDB_1lox-17Puro-5: 0.25, iPS_cells:PDB_1lox-17Puro-10: 0.25, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.25, iPS_cells:PDB_2lox-22: 0.25, iPS_cells:PDB_1lox-21Puro-20: 0.25, iPS_cells:PDB_1lox-21Puro-26: 0.25, iPS_cells:PDB_2lox-5: 0.25 |
| T200_GTATTTCAGGCTCTAT-1 | Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys | 0.15 | 428.08 | Raw ScoresMacrophage:monocyte-derived:M-CSF/IFNg: 0.39, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.38, DC:monocyte-derived:LPS: 0.38, Macrophage:monocyte-derived:IFNa: 0.38, DC:monocyte-derived:AEC-conditioned: 0.38, DC:monocyte-derived:Poly(IC): 0.38, Macrophage:monocyte-derived:M-CSF: 0.38, DC:monocyte-derived:anti-DC-SIGN_2h: 0.38, Macrophage:monocyte-derived:S._aureus: 0.38, DC:monocyte-derived:Galectin-1: 0.38 |
| T200_CTCTCAGAGCCACCGT-1 | Neurons:adrenal_medulla_cell_line | 0.19 | 408.68 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.39, Neuroepithelial_cell:ESC-derived: 0.34, Astrocyte:Embryonic_stem_cell-derived: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.31, iPS_cells:PDB_1lox-17Puro-5: 0.31, iPS_cells:PDB_1lox-21Puro-26: 0.31, iPS_cells:PDB_1lox-21Puro-20: 0.31, iPS_cells:PDB_2lox-22: 0.31, iPS_cells:PDB_2lox-21: 0.31, Neurons:ES_cell-derived_neural_precursor: 0.31 |
| T200_TTGTTCAGTGCAAGAC-1 | Neurons:adrenal_medulla_cell_line | 0.19 | 407.14 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.39, Neuroepithelial_cell:ESC-derived: 0.36, iPS_cells:PDB_1lox-17Puro-10: 0.35, iPS_cells:PDB_1lox-21Puro-26: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.35, Embryonic_stem_cells: 0.34, iPS_cells:PDB_2lox-22: 0.34, iPS_cells:PDB_2lox-17: 0.34, iPS_cells:PDB_2lox-21: 0.34 |
| T200_TCTTGCGAGAAACCCG-1 | Fibroblasts:foreskin | 0.13 | 406.80 | Raw ScoresMSC: 0.35, Neuroepithelial_cell:ESC-derived: 0.34, Fibroblasts:foreskin: 0.34, Neurons:Schwann_cell: 0.34, Fibroblasts:breast: 0.34, iPS_cells:CRL2097_foreskin: 0.33, Tissue_stem_cells:BM_MSC: 0.33, Smooth_muscle_cells:vascular: 0.33, Neurons:adrenal_medulla_cell_line: 0.33, Astrocyte:Embryonic_stem_cell-derived: 0.33 |
| T200_TATTGGGCACGACGAA-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 403.05 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.34, Neuroepithelial_cell:ESC-derived: 0.3, Astrocyte:Embryonic_stem_cell-derived: 0.3, iPS_cells:PDB_1lox-17Puro-5: 0.27, iPS_cells:PDB_1lox-17Puro-10: 0.27, iPS_cells:PDB_1lox-21Puro-26: 0.27, Neurons:ES_cell-derived_neural_precursor: 0.27, iPS_cells:PDB_1lox-21Puro-20: 0.27, iPS_cells:PDB_2lox-22: 0.27, iPS_cells:PDB_2lox-17: 0.26 |
| T200_ATCAGGTAGACTTAAG-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 402.35 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.32, Neuroepithelial_cell:ESC-derived: 0.29, Astrocyte:Embryonic_stem_cell-derived: 0.28, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.26, iPS_cells:PDB_1lox-17Puro-10: 0.26, iPS_cells:PDB_1lox-21Puro-20: 0.26, Embryonic_stem_cells: 0.26, iPS_cells:PDB_1lox-17Puro-5: 0.26, iPS_cells:PDB_2lox-22: 0.26, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.26 |
| T200_ACGGTCGCACATTCTT-1 | Neurons:adrenal_medulla_cell_line | 0.12 | 399.30 | Raw ScoresFibroblasts:breast: 0.32, Tissue_stem_cells:BM_MSC:BMP2: 0.32, Smooth_muscle_cells:bronchial: 0.32, Tissue_stem_cells:BM_MSC:TGFb3: 0.32, iPS_cells:CRL2097_foreskin: 0.32, Smooth_muscle_cells:vascular: 0.31, Smooth_muscle_cells:bronchial:vit_D: 0.31, iPS_cells:adipose_stem_cells: 0.31, MSC: 0.31, Chondrocytes:MSC-derived: 0.31 |
| T200_AGGGTCCCAGGAACCA-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 392.44 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Astrocyte:Embryonic_stem_cell-derived: 0.31, Neuroepithelial_cell:ESC-derived: 0.3, Embryonic_stem_cells: 0.27, Tissue_stem_cells:dental_pulp: 0.26, Fibroblasts:breast: 0.26, Smooth_muscle_cells:umbilical_vein: 0.26, Neurons:ES_cell-derived_neural_precursor: 0.26, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.26, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.26 |
| T200_TCCTCCCAGCCTCATA-1 | Neurons:adrenal_medulla_cell_line | 0.17 | 385.68 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.35, Neuroepithelial_cell:ESC-derived: 0.31, Astrocyte:Embryonic_stem_cell-derived: 0.3, iPS_cells:PDB_1lox-21Puro-20: 0.27, Neurons:ES_cell-derived_neural_precursor: 0.27, iPS_cells:PDB_1lox-17Puro-10: 0.27, iPS_cells:PDB_1lox-21Puro-26: 0.27, iPS_cells:PDB_1lox-17Puro-5: 0.27, iPS_cells:PDB_2lox-22: 0.27, Embryonic_stem_cells: 0.27 |
| T200_CCTTGTGAGTTACGGG-1 | DC:monocyte-derived:AEC-conditioned | 0.12 | 383.02 | Raw ScoresMacrophage:monocyte-derived:IFNa: 0.36, DC:monocyte-derived:AEC-conditioned: 0.36, Macrophage:monocyte-derived:S._aureus: 0.36, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.35, DC:monocyte-derived:anti-DC-SIGN_2h: 0.35, Macrophage:monocyte-derived:M-CSF/IFNg: 0.35, Macrophage:Alveolar:B._anthacis_spores: 0.35, DC:monocyte-derived:LPS: 0.35, Macrophage:monocyte-derived:M-CSF: 0.35, Macrophage:Alveolar: 0.35 |
| T200_GAACTGTAGTACAACA-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 382.37 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.36, Astrocyte:Embryonic_stem_cell-derived: 0.32, Neuroepithelial_cell:ESC-derived: 0.31, iPS_cells:PDB_1lox-17Puro-10: 0.29, iPS_cells:PDB_1lox-21Puro-20: 0.29, iPS_cells:PDB_1lox-17Puro-5: 0.29, iPS_cells:PDB_1lox-21Puro-26: 0.29, iPS_cells:skin_fibroblast-derived: 0.28, Embryonic_stem_cells: 0.28, iPS_cells:PDB_2lox-22: 0.28 |
| T200_GTAGAGGGTGATTGGG-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 380.96 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.34, Neuroepithelial_cell:ESC-derived: 0.3, Astrocyte:Embryonic_stem_cell-derived: 0.3, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.28, iPS_cells:PDB_1lox-17Puro-5: 0.27, iPS_cells:PDB_1lox-21Puro-20: 0.27, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.27, iPS_cells:adipose_stem_cell-derived:lentiviral: 0.27, iPS_cells:PDB_2lox-22: 0.27, iPS_cells:PDB_2lox-17: 0.27 |
| T200_TCATTACAGGGAGGAC-1 | Neurons:adrenal_medulla_cell_line | 0.18 | 380.55 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.34, Neuroepithelial_cell:ESC-derived: 0.29, Astrocyte:Embryonic_stem_cell-derived: 0.28, iPS_cells:PDB_1lox-21Puro-20: 0.27, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.27, iPS_cells:PDB_2lox-22: 0.27, iPS_cells:PDB_1lox-17Puro-5: 0.27, iPS_cells:PDB_1lox-17Puro-10: 0.27, iPS_cells:PDB_1lox-21Puro-26: 0.27, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.27 |
| T200_CATGAGTGTTACACAC-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 380.37 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Neuroepithelial_cell:ESC-derived: 0.31, Astrocyte:Embryonic_stem_cell-derived: 0.3, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.28, iPS_cells:PDB_2lox-22: 0.28, Neurons:ES_cell-derived_neural_precursor: 0.28, iPS_cells:PDB_1lox-17Puro-10: 0.27, iPS_cells:PDB_1lox-17Puro-5: 0.27, iPS_cells:PDB_1lox-21Puro-20: 0.27, iPS_cells:PDB_1lox-21Puro-26: 0.27 |
| T200_ATCTTCACACAGCCTG-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 367.82 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Neuroepithelial_cell:ESC-derived: 0.3, Astrocyte:Embryonic_stem_cell-derived: 0.3, iPS_cells:PDB_1lox-17Puro-10: 0.28, iPS_cells:PDB_1lox-17Puro-5: 0.28, iPS_cells:PDB_1lox-21Puro-20: 0.28, iPS_cells:PDB_1lox-21Puro-26: 0.28, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.28, iPS_cells:PDB_2lox-22: 0.28, iPS_cells:PDB_2lox-21: 0.28 |
| T200_CCCTAACGTTGTGTTG-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 366.28 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.32, Astrocyte:Embryonic_stem_cell-derived: 0.29, Neuroepithelial_cell:ESC-derived: 0.28, iPS_cells:PDB_1lox-21Puro-20: 0.26, iPS_cells:PDB_1lox-17Puro-5: 0.26, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.26, iPS_cells:PDB_1lox-17Puro-10: 0.26, Embryonic_stem_cells: 0.26, iPS_cells:PDB_1lox-21Puro-26: 0.26, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.26 |
| T200_TGGGAGACACGACGAA-1 | Neurons:adrenal_medulla_cell_line | 0.14 | 364.05 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.31, Astrocyte:Embryonic_stem_cell-derived: 0.28, Neuroepithelial_cell:ESC-derived: 0.28, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.26, iPS_cells:PDB_1lox-17Puro-5: 0.26, iPS_cells:PDB_1lox-21Puro-20: 0.26, iPS_cells:PDB_1lox-17Puro-10: 0.26, Neurons:ES_cell-derived_neural_precursor: 0.25, iPS_cells:PDB_1lox-21Puro-26: 0.25, iPS_cells:PDB_2lox-22: 0.25 |
| T200_GATAGAAAGACATCCT-1 | Neurons:adrenal_medulla_cell_line | 0.16 | 362.83 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.32, Astrocyte:Embryonic_stem_cell-derived: 0.29, Neuroepithelial_cell:ESC-derived: 0.28, Neurons:ES_cell-derived_neural_precursor: 0.25, Embryonic_stem_cells: 0.25, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.24, iPS_cells:PDB_1lox-17Puro-5: 0.24, iPS_cells:PDB_1lox-21Puro-20: 0.24, iPS_cells:PDB_1lox-17Puro-10: 0.24, iPS_cells:PDB_1lox-21Puro-26: 0.24 |
| T200_CAATCGACAAGGGTCA-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 359.85 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Neuroepithelial_cell:ESC-derived: 0.3, Astrocyte:Embryonic_stem_cell-derived: 0.3, iPS_cells:PDB_1lox-21Puro-20: 0.29, iPS_cells:PDB_1lox-17Puro-5: 0.29, iPS_cells:PDB_1lox-21Puro-26: 0.29, iPS_cells:PDB_2lox-22: 0.29, iPS_cells:PDB_1lox-17Puro-10: 0.29, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.29, iPS_cells:PDB_2lox-17: 0.29 |
| T200_CCGGTAGTCATAGACC-1 | Macrophage:monocyte-derived:S._aureus | 0.09 | 357.63 | Raw ScoresMacrophage:monocyte-derived:S._aureus: 0.3, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.3, Macrophage:monocyte-derived:IFNa: 0.3, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.3, DC:monocyte-derived:LPS: 0.3, Monocyte:CD16-: 0.3, DC:monocyte-derived:anti-DC-SIGN_2h: 0.3, DC:monocyte-derived:Galectin-1: 0.3, DC:monocyte-derived:AEC-conditioned: 0.3, Macrophage:monocyte-derived:M-CSF/IFNg: 0.3 |
| T200_GCACTAATCACATCAG-1 | Neurons:adrenal_medulla_cell_line | 0.13 | 356.89 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Astrocyte:Embryonic_stem_cell-derived: 0.26, Neuroepithelial_cell:ESC-derived: 0.26, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.23, iPS_cells:PDB_1lox-21Puro-20: 0.23, iPS_cells:PDB_1lox-17Puro-5: 0.23, Fibroblasts:breast: 0.23, iPS_cells:PDB_2lox-22: 0.23, iPS_cells:PDB_1lox-17Puro-10: 0.23, iPS_cells:PDB_1lox-21Puro-26: 0.23 |
| T200_CTACCCAGTTTGGAAA-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 356.59 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.31, Astrocyte:Embryonic_stem_cell-derived: 0.29, Neuroepithelial_cell:ESC-derived: 0.27, iPS_cells:PDB_1lox-21Puro-20: 0.24, Neurons:ES_cell-derived_neural_precursor: 0.24, iPS_cells:PDB_1lox-17Puro-5: 0.24, iPS_cells:PDB_1lox-17Puro-10: 0.24, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.24, iPS_cells:PDB_1lox-21Puro-26: 0.23, iPS_cells:PDB_2lox-21: 0.23 |
| T200_CACAGGCCAACAAGTA-1 | Neurons:adrenal_medulla_cell_line | 0.15 | 344.38 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.32, Astrocyte:Embryonic_stem_cell-derived: 0.29, Neuroepithelial_cell:ESC-derived: 0.28, iPS_cells:PDB_1lox-21Puro-20: 0.26, iPS_cells:PDB_2lox-22: 0.26, iPS_cells:PDB_2lox-21: 0.26, iPS_cells:PDB_1lox-17Puro-5: 0.26, iPS_cells:PDB_1lox-17Puro-10: 0.26, iPS_cells:PDB_1lox-21Puro-26: 0.26, iPS_cells:PDB_2lox-17: 0.26 |
Below shows the significant enrichments of this GEP for literature curated gene lists
This data was procured from existing single cell RNA-seq maps of neuroblastoma or related relevant data.
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by the P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
N Chromafin (Hanemaaijer)
Marker genes obtained from Supplementary Table SD of Hanemaaijer et al (PMID 33500353). The authors generated single-cell RNA-seq data (sort-seq, 2,229 cells total) from mouse adrenal glads at E13.5, E14.5, E17.5, E18.5, P1 and P5. These were marker genes that matched with a similar dataset generated by Furlan et al (PMID 28684471). This particular set of markers are for the Norepinepherine Chromaffin subcluster, which is part of the Adrenal Medulla cluster.:
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.51e-05
Mean rank of genes in gene set: 2499.19
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| NAP1L5 | 0.0040741 | 42 | GTEx | DepMap | Descartes | 1.66 | 135.46 |
| SYN2 | 0.0032153 | 69 | GTEx | DepMap | Descartes | 0.27 | 11.04 |
| CELF4 | 0.0029256 | 89 | GTEx | DepMap | Descartes | 3.47 | 133.65 |
| SCG5 | 0.0028867 | 91 | GTEx | DepMap | Descartes | 5.31 | 644.91 |
| CACNA2D1 | 0.0023589 | 135 | GTEx | DepMap | Descartes | 1.12 | 22.99 |
| SCG3 | 0.0020511 | 180 | GTEx | DepMap | Descartes | 4.14 | 197.86 |
| LGR5 | 0.0014562 | 323 | GTEx | DepMap | Descartes | 0.10 | 3.12 |
| SNAP25 | 0.0008626 | 741 | GTEx | DepMap | Descartes | 1.58 | 104.68 |
| GNAS | 0.0006215 | 1112 | GTEx | DepMap | Descartes | 32.91 | 1225.20 |
| PTPRN | 0.0004796 | 1434 | GTEx | DepMap | Descartes | 0.65 | 19.78 |
| PCLO | 0.0003679 | 1735 | GTEx | DepMap | Descartes | 0.97 | 7.20 |
| ADCYAP1R1 | 0.0001980 | 2502 | GTEx | DepMap | Descartes | 0.16 | 3.42 |
| CXCL14 | 0.0001039 | 3110 | GTEx | DepMap | Descartes | 0.02 | 1.97 |
| PPFIA2 | 0.0000639 | 3438 | GTEx | DepMap | Descartes | 0.21 | 5.97 |
| SLCO3A1 | -0.0013168 | 12450 | GTEx | DepMap | Descartes | 0.40 | 10.54 |
| C1QL1 | -0.0018901 | 12536 | GTEx | DepMap | Descartes | 0.27 | 26.61 |
Sympathoblasts (Kildisiute)
Sympathoblasts markers obtained from Kildisiute et al, Supplmenentary Table 2, references supporting these genes are provided in Supp Table S2 of Kildisiute et al (PMID 33547074) https://www.science.org/doi/suppl/10.1126/sciadv.abd3311/suppl_file/abd3311_tables_s1_to_s12.xlsx:
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.06e-03
Mean rank of genes in gene set: 274
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| NPY | 0.0062200 | 15 | GTEx | DepMap | Descartes | 54.84 | 11736.49 |
| GAP43 | 0.0021549 | 162 | GTEx | DepMap | Descartes | 5.78 | 449.57 |
| BCL2 | 0.0009714 | 645 | GTEx | DepMap | Descartes | 0.80 | 16.62 |
IFN Response (Kinker)
These marker genes were obtained in a pan cancer cell lines NMF analysis of scRNA-eq data in Kinker et al (PMID 33128048) - 10 metaprograms were recovered that manifest across cancers, this program contained interferon response genes.:
Wilcoxon ranksum test P-value for gene set overrepresentation: 4.50e-03
Mean rank of genes in gene set: 2044.4
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| IFIT1 | 0.0119898 | 1 | GTEx | DepMap | Descartes | 3.74 | 125.06 |
| IFIT3 | 0.0109563 | 3 | GTEx | DepMap | Descartes | 2.88 | 178.74 |
| IFIT2 | 0.0023667 | 134 | GTEx | DepMap | Descartes | 0.52 | 21.74 |
| ISG15 | 0.0018666 | 207 | GTEx | DepMap | Descartes | 7.97 | 1510.77 |
| ISG20 | -0.0004166 | 9877 | GTEx | DepMap | Descartes | 0.15 | 3.27 |
Below shows ranks on this GEP for literature curated gene lists for large gene sets
These include those reported as mesenchymal/adrenergic by Van Groningen et al.
High ranks indicate this gene is a driver of this GEP (note these results are not ordered).
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
VanGroningen Adrenergic Genes
Adrenergic marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 5.67e-02
Mean rank of genes in gene set: 5950.58
Median rank of genes in gene set: 5457
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| FBXO8 | 0.0062642 | 14 | GTEx | DepMap | Descartes | 1.39 | 106.09 |
| NPY | 0.0062200 | 15 | GTEx | DepMap | Descartes | 54.84 | 11736.49 |
| NNAT | 0.0051197 | 22 | GTEx | DepMap | Descartes | 7.58 | 911.70 |
| RTN1 | 0.0050951 | 23 | GTEx | DepMap | Descartes | 16.39 | 763.13 |
| TMEM108 | 0.0041530 | 39 | GTEx | DepMap | Descartes | 0.83 | 25.59 |
| NAP1L5 | 0.0040741 | 42 | GTEx | DepMap | Descartes | 1.66 | 135.46 |
| MMD | 0.0038208 | 47 | GTEx | DepMap | Descartes | 2.45 | 140.40 |
| TUBB2A | 0.0036454 | 52 | GTEx | DepMap | Descartes | 8.99 | 803.41 |
| PRSS12 | 0.0034987 | 57 | GTEx | DepMap | Descartes | 0.99 | 32.82 |
| CEP44 | 0.0032119 | 70 | GTEx | DepMap | Descartes | 1.61 | 58.25 |
| RPS6KA2 | 0.0030018 | 85 | GTEx | DepMap | Descartes | 0.67 | 16.97 |
| RBMS3 | 0.0029939 | 86 | GTEx | DepMap | Descartes | 4.12 | 74.73 |
| MAP2 | 0.0028708 | 92 | GTEx | DepMap | Descartes | 3.16 | 47.59 |
| SCN3A | 0.0023221 | 136 | GTEx | DepMap | Descartes | 1.15 | 19.40 |
| OLFM1 | 0.0023203 | 139 | GTEx | DepMap | Descartes | 1.49 | 83.10 |
| NRSN1 | 0.0021778 | 155 | GTEx | DepMap | Descartes | 1.66 | 106.27 |
| PKIA | 0.0021587 | 161 | GTEx | DepMap | Descartes | 1.86 | 71.09 |
| GAP43 | 0.0021549 | 162 | GTEx | DepMap | Descartes | 5.78 | 449.57 |
| OLA1 | 0.0021071 | 169 | GTEx | DepMap | Descartes | 4.12 | 138.30 |
| FKBP1B | 0.0021063 | 171 | GTEx | DepMap | Descartes | 1.38 | 130.86 |
| DPYSL2 | 0.0020939 | 175 | GTEx | DepMap | Descartes | 3.76 | 117.18 |
| SCG3 | 0.0020511 | 180 | GTEx | DepMap | Descartes | 4.14 | 197.86 |
| FAM171B | 0.0020057 | 181 | GTEx | DepMap | Descartes | 1.02 | 27.36 |
| SEC11C | 0.0020037 | 182 | GTEx | DepMap | Descartes | 3.26 | 229.05 |
| ELAVL4 | 0.0019962 | 183 | GTEx | DepMap | Descartes | 9.24 | 345.12 |
| MYRIP | 0.0019057 | 200 | GTEx | DepMap | Descartes | 0.11 | 3.02 |
| CCNI | 0.0018666 | 208 | GTEx | DepMap | Descartes | 21.93 | 1167.22 |
| INA | 0.0018604 | 211 | GTEx | DepMap | Descartes | 2.71 | 123.84 |
| EIF1B | 0.0018336 | 220 | GTEx | DepMap | Descartes | 5.45 | 845.01 |
| ELAVL2 | 0.0018003 | 226 | GTEx | DepMap | Descartes | 2.01 | 75.60 |
VanGroningen Mesenchymal Genes
Mesenchymal marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 7387.25
Median rank of genes in gene set: 8052
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| STAT1 | 0.0083173 | 6 | GTEx | DepMap | Descartes | 3.98 | 141.58 |
| PLSCR1 | 0.0034636 | 58 | GTEx | DepMap | Descartes | 1.22 | 92.00 |
| ATP1B1 | 0.0031777 | 72 | GTEx | DepMap | Descartes | 3.72 | 220.56 |
| IGFBP5 | 0.0027284 | 100 | GTEx | DepMap | Descartes | 4.70 | 123.34 |
| PTN | 0.0019916 | 186 | GTEx | DepMap | Descartes | 1.25 | 124.37 |
| CRABP2 | 0.0019325 | 195 | GTEx | DepMap | Descartes | 0.49 | 69.89 |
| FGFR1 | 0.0019249 | 198 | GTEx | DepMap | Descartes | 0.52 | 12.19 |
| DKK3 | 0.0016123 | 269 | GTEx | DepMap | Descartes | 0.60 | 9.96 |
| COL11A1 | 0.0015231 | 295 | GTEx | DepMap | Descartes | 0.17 | 3.66 |
| BMP5 | 0.0014576 | 321 | GTEx | DepMap | Descartes | 0.03 | 0.98 |
| PDE3A | 0.0013906 | 357 | GTEx | DepMap | Descartes | 0.25 | 4.87 |
| BNC2 | 0.0013454 | 380 | GTEx | DepMap | Descartes | 0.09 | 0.96 |
| CRTAP | 0.0013398 | 385 | GTEx | DepMap | Descartes | 0.76 | 15.97 |
| PPIB | 0.0012015 | 456 | GTEx | DepMap | Descartes | 6.91 | 844.81 |
| SVIL | 0.0011276 | 507 | GTEx | DepMap | Descartes | 0.27 | 4.89 |
| WNT5A | 0.0010581 | 553 | GTEx | DepMap | Descartes | 0.05 | 1.13 |
| SCRG1 | 0.0010468 | 565 | GTEx | DepMap | Descartes | 0.40 | 15.39 |
| EGR3 | 0.0010302 | 574 | GTEx | DepMap | Descartes | 0.69 | 22.40 |
| DMD | 0.0010291 | 576 | GTEx | DepMap | Descartes | 0.39 | 4.39 |
| KDM5B | 0.0009823 | 628 | GTEx | DepMap | Descartes | 1.18 | 18.73 |
| STAT3 | 0.0009739 | 642 | GTEx | DepMap | Descartes | 1.75 | 52.17 |
| APP | 0.0009397 | 670 | GTEx | DepMap | Descartes | 1.81 | 78.11 |
| GPC6 | 0.0008748 | 729 | GTEx | DepMap | Descartes | 0.19 | 3.95 |
| IL6ST | 0.0008215 | 790 | GTEx | DepMap | Descartes | 0.44 | 7.09 |
| ATXN1 | 0.0007971 | 818 | GTEx | DepMap | Descartes | 0.49 | 7.35 |
| EXTL2 | 0.0007926 | 826 | GTEx | DepMap | Descartes | 0.53 | 24.76 |
| ATP2B4 | 0.0007762 | 849 | GTEx | DepMap | Descartes | 0.25 | 4.40 |
| ARMCX2 | 0.0007538 | 881 | GTEx | DepMap | Descartes | 0.41 | 20.85 |
| MYL12B | 0.0007527 | 884 | GTEx | DepMap | Descartes | 4.29 | 501.65 |
| GNAI1 | 0.0007494 | 889 | GTEx | DepMap | Descartes | 0.73 | 11.30 |
Descartes adrenocortical markers
Top 50 marker genes of adrenocortical cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/adrenocortical/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.47e-01
Mean rank of genes in gene set: 5862.42
Median rank of genes in gene set: 5701.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| GSTA4 | 0.0018219 | 223 | GTEx | DepMap | Descartes | 2.11 | 184.77 |
| SH3BP5 | 0.0015813 | 279 | GTEx | DepMap | Descartes | 1.09 | 48.11 |
| TM7SF2 | 0.0011521 | 490 | GTEx | DepMap | Descartes | 0.81 | 59.02 |
| SGCZ | 0.0010864 | 535 | GTEx | DepMap | Descartes | 0.06 | 1.18 |
| MSMO1 | 0.0006002 | 1160 | GTEx | DepMap | Descartes | 0.52 | 33.74 |
| POR | 0.0005612 | 1236 | GTEx | DepMap | Descartes | 0.52 | 29.99 |
| CLU | 0.0004922 | 1394 | GTEx | DepMap | Descartes | 1.82 | 104.67 |
| GRAMD1B | 0.0004139 | 1589 | GTEx | DepMap | Descartes | 0.12 | 2.30 |
| PEG3 | 0.0003098 | 1961 | GTEx | DepMap | Descartes | 0.54 | NA |
| PDE10A | 0.0003028 | 1996 | GTEx | DepMap | Descartes | 0.18 | 3.27 |
| JAKMIP2 | 0.0001876 | 2556 | GTEx | DepMap | Descartes | 0.55 | 8.93 |
| SH3PXD2B | 0.0000640 | 3436 | GTEx | DepMap | Descartes | 0.10 | 1.85 |
| HMGCS1 | 0.0000350 | 3709 | GTEx | DepMap | Descartes | 0.35 | 9.15 |
| HMGCR | 0.0000317 | 3744 | GTEx | DepMap | Descartes | 0.36 | 11.67 |
| DHCR7 | -0.0000008 | 4112 | GTEx | DepMap | Descartes | 0.15 | 8.14 |
| FDXR | -0.0000480 | 4757 | GTEx | DepMap | Descartes | 0.20 | 11.30 |
| INHA | -0.0000513 | 4809 | GTEx | DepMap | Descartes | 0.01 | 0.45 |
| BAIAP2L1 | -0.0000789 | 5236 | GTEx | DepMap | Descartes | 0.01 | 0.44 |
| FREM2 | -0.0001357 | 6167 | GTEx | DepMap | Descartes | 0.00 | 0.01 |
| STAR | -0.0001374 | 6192 | GTEx | DepMap | Descartes | 0.02 | 0.82 |
| ERN1 | -0.0001496 | 6396 | GTEx | DepMap | Descartes | 0.11 | 2.27 |
| FDPS | -0.0001893 | 6964 | GTEx | DepMap | Descartes | 1.26 | 85.27 |
| LDLR | -0.0002234 | 7465 | GTEx | DepMap | Descartes | 0.06 | 1.36 |
| SCAP | -0.0002390 | 7670 | GTEx | DepMap | Descartes | 0.27 | 8.95 |
| DHCR24 | -0.0003022 | 8508 | GTEx | DepMap | Descartes | 0.09 | 1.93 |
| CYB5B | -0.0004006 | 9711 | GTEx | DepMap | Descartes | 0.37 | 11.84 |
| NPC1 | -0.0004496 | 10190 | GTEx | DepMap | Descartes | 0.05 | 1.83 |
| SLC16A9 | -0.0004503 | 10195 | GTEx | DepMap | Descartes | 0.10 | 3.20 |
| IGF1R | -0.0004511 | 10200 | GTEx | DepMap | Descartes | 0.37 | 4.33 |
| FDX1 | -0.0004701 | 10350 | GTEx | DepMap | Descartes | 0.62 | 27.74 |
Descartes chromaffin markers
Top 50 marker genes of chromaffin cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/chromaffin/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.47e-01
Mean rank of genes in gene set: 5887.98
Median rank of genes in gene set: 4168
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| NPY | 0.0062200 | 15 | GTEx | DepMap | Descartes | 54.84 | 11736.49 |
| MAB21L2 | 0.0048588 | 29 | GTEx | DepMap | Descartes | 3.11 | 188.54 |
| TUBB2A | 0.0036454 | 52 | GTEx | DepMap | Descartes | 8.99 | 803.41 |
| RYR2 | 0.0022885 | 143 | GTEx | DepMap | Descartes | 0.41 | 3.70 |
| GAP43 | 0.0021549 | 162 | GTEx | DepMap | Descartes | 5.78 | 449.57 |
| MAB21L1 | 0.0019871 | 188 | GTEx | DepMap | Descartes | 2.84 | 154.45 |
| ELAVL2 | 0.0018003 | 226 | GTEx | DepMap | Descartes | 2.01 | 75.60 |
| TUBB2B | 0.0016726 | 256 | GTEx | DepMap | Descartes | 30.78 | 2424.10 |
| TUBA1A | 0.0013997 | 352 | GTEx | DepMap | Descartes | 46.96 | 3569.15 |
| EYA1 | 0.0012056 | 451 | GTEx | DepMap | Descartes | 0.35 | 13.20 |
| MAP1B | 0.0009455 | 662 | GTEx | DepMap | Descartes | 13.78 | 170.38 |
| SLC44A5 | 0.0009437 | 663 | GTEx | DepMap | Descartes | 0.14 | 4.61 |
| REEP1 | 0.0009075 | 702 | GTEx | DepMap | Descartes | 0.75 | 27.63 |
| SYNPO2 | 0.0008882 | 716 | GTEx | DepMap | Descartes | 0.63 | 6.40 |
| BASP1 | 0.0008408 | 768 | GTEx | DepMap | Descartes | 13.87 | 1134.98 |
| MARCH11 | 0.0006678 | 1023 | GTEx | DepMap | Descartes | 4.71 | NA |
| RPH3A | 0.0004849 | 1421 | GTEx | DepMap | Descartes | 0.14 | 4.06 |
| EPHA6 | 0.0002680 | 2147 | GTEx | DepMap | Descartes | 0.06 | 2.00 |
| MLLT11 | 0.0002387 | 2274 | GTEx | DepMap | Descartes | 8.29 | 492.00 |
| CNKSR2 | 0.0000420 | 3636 | GTEx | DepMap | Descartes | 0.19 | 3.65 |
| TMEFF2 | -0.0000054 | 4168 | GTEx | DepMap | Descartes | 0.41 | 19.27 |
| PTCHD1 | -0.0000131 | 4262 | GTEx | DepMap | Descartes | 0.13 | 1.73 |
| GAL | -0.0002483 | 7784 | GTEx | DepMap | Descartes | 1.83 | 371.35 |
| CNTFR | -0.0003069 | 8577 | GTEx | DepMap | Descartes | 0.76 | 58.95 |
| HS3ST5 | -0.0003500 | 9127 | GTEx | DepMap | Descartes | 0.03 | 1.26 |
| ANKFN1 | -0.0004894 | 10523 | GTEx | DepMap | Descartes | 0.01 | 0.15 |
| ALK | -0.0004980 | 10577 | GTEx | DepMap | Descartes | 0.04 | 1.09 |
| GREM1 | -0.0005973 | 11234 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| KCNB2 | -0.0006958 | 11631 | GTEx | DepMap | Descartes | 0.07 | 3.07 |
| STMN2 | -0.0007238 | 11724 | GTEx | DepMap | Descartes | 22.60 | 1805.85 |
Descartes Vascular_endothelial markers
Top 50 marker genes of Vascular_endothelial cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/vascular_endothelial/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 7.07e-01
Mean rank of genes in gene set: 6595.13
Median rank of genes in gene set: 7727.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| MYRIP | 0.0019057 | 200 | GTEx | DepMap | Descartes | 0.11 | 3.02 |
| ARHGAP29 | 0.0016032 | 271 | GTEx | DepMap | Descartes | 0.71 | 11.88 |
| CHRM3 | 0.0013316 | 387 | GTEx | DepMap | Descartes | 0.55 | 9.45 |
| RAMP2 | 0.0007993 | 814 | GTEx | DepMap | Descartes | 1.92 | 328.74 |
| GALNT15 | 0.0004101 | 1598 | GTEx | DepMap | Descartes | 0.03 | NA |
| NOTCH4 | 0.0003897 | 1665 | GTEx | DepMap | Descartes | 0.22 | 4.90 |
| EHD3 | 0.0002675 | 2148 | GTEx | DepMap | Descartes | 0.05 | 1.59 |
| F8 | 0.0002419 | 2258 | GTEx | DepMap | Descartes | 0.03 | 0.55 |
| CLDN5 | 0.0000446 | 3608 | GTEx | DepMap | Descartes | 0.15 | 8.22 |
| ID1 | 0.0000252 | 3806 | GTEx | DepMap | Descartes | 0.28 | 35.13 |
| SHANK3 | 0.0000104 | 3974 | GTEx | DepMap | Descartes | 0.04 | 0.78 |
| NR5A2 | -0.0000280 | 4460 | GTEx | DepMap | Descartes | 0.00 | 0.05 |
| TEK | -0.0001645 | 6624 | GTEx | DepMap | Descartes | 0.00 | 0.02 |
| HYAL2 | -0.0001730 | 6736 | GTEx | DepMap | Descartes | 0.69 | 24.90 |
| SHE | -0.0002102 | 7268 | GTEx | DepMap | Descartes | 0.01 | 0.13 |
| RASIP1 | -0.0002207 | 7424 | GTEx | DepMap | Descartes | 0.01 | 0.41 |
| KANK3 | -0.0002295 | 7549 | GTEx | DepMap | Descartes | 0.01 | 0.65 |
| ESM1 | -0.0002349 | 7611 | GTEx | DepMap | Descartes | 0.01 | 0.27 |
| ROBO4 | -0.0002422 | 7708 | GTEx | DepMap | Descartes | 0.02 | 0.39 |
| NPR1 | -0.0002452 | 7747 | GTEx | DepMap | Descartes | 0.00 | 0.04 |
| CDH5 | -0.0002529 | 7854 | GTEx | DepMap | Descartes | 0.02 | 0.49 |
| CRHBP | -0.0002555 | 7886 | GTEx | DepMap | Descartes | 0.01 | 0.89 |
| KDR | -0.0002615 | 7978 | GTEx | DepMap | Descartes | 0.02 | 0.48 |
| BTNL9 | -0.0002772 | 8186 | GTEx | DepMap | Descartes | 0.00 | 0.02 |
| CEACAM1 | -0.0002860 | 8293 | GTEx | DepMap | Descartes | 0.00 | 0.11 |
| FLT4 | -0.0002890 | 8326 | GTEx | DepMap | Descartes | 0.01 | 0.10 |
| PTPRB | -0.0002913 | 8363 | GTEx | DepMap | Descartes | 0.02 | 0.16 |
| CALCRL | -0.0003204 | 8757 | GTEx | DepMap | Descartes | 0.02 | 0.28 |
| MMRN2 | -0.0003243 | 8807 | GTEx | DepMap | Descartes | 0.02 | 0.40 |
| PLVAP | -0.0003360 | 8965 | GTEx | DepMap | Descartes | 0.07 | 3.20 |
Descartes stromal markers
Top 50 marker genes of stromal cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/stromal/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 5.98e-01
Mean rank of genes in gene set: 6409.84
Median rank of genes in gene set: 6182
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| PCOLCE | 0.0018892 | 203 | GTEx | DepMap | Descartes | 2.95 | 284.24 |
| CLDN11 | 0.0013128 | 402 | GTEx | DepMap | Descartes | 0.16 | 9.13 |
| FREM1 | 0.0007934 | 823 | GTEx | DepMap | Descartes | 0.05 | 0.64 |
| DKK2 | 0.0007900 | 829 | GTEx | DepMap | Descartes | 0.03 | 1.36 |
| ADAMTSL3 | 0.0002885 | 2040 | GTEx | DepMap | Descartes | 0.02 | 0.31 |
| EDNRA | 0.0002502 | 2217 | GTEx | DepMap | Descartes | 0.08 | 2.41 |
| PRICKLE1 | 0.0002333 | 2297 | GTEx | DepMap | Descartes | 0.31 | 7.64 |
| LRRC17 | 0.0001541 | 2761 | GTEx | DepMap | Descartes | 0.03 | 2.18 |
| LAMC3 | 0.0000495 | 3568 | GTEx | DepMap | Descartes | 0.01 | 0.10 |
| GLI2 | -0.0000094 | 4211 | GTEx | DepMap | Descartes | 0.00 | 0.06 |
| PCDH18 | -0.0000480 | 4756 | GTEx | DepMap | Descartes | 0.04 | 1.22 |
| RSPO3 | -0.0000617 | 4996 | GTEx | DepMap | Descartes | 0.00 | NA |
| GAS2 | -0.0000656 | 5056 | GTEx | DepMap | Descartes | 0.01 | 0.67 |
| CCDC80 | -0.0000873 | 5377 | GTEx | DepMap | Descartes | 0.06 | 0.78 |
| ITGA11 | -0.0000951 | 5505 | GTEx | DepMap | Descartes | 0.00 | 0.10 |
| POSTN | -0.0001085 | 5710 | GTEx | DepMap | Descartes | 0.04 | 1.57 |
| IGFBP3 | -0.0001142 | 5810 | GTEx | DepMap | Descartes | 0.02 | 1.01 |
| ISLR | -0.0001191 | 5893 | GTEx | DepMap | Descartes | 0.01 | 0.94 |
| SFRP2 | -0.0001215 | 5940 | GTEx | DepMap | Descartes | 0.00 | 0.07 |
| BICC1 | -0.0001242 | 5981 | GTEx | DepMap | Descartes | 0.03 | 0.71 |
| COL6A3 | -0.0001325 | 6107 | GTEx | DepMap | Descartes | 0.16 | 2.17 |
| LOX | -0.0001339 | 6134 | GTEx | DepMap | Descartes | 0.00 | 0.06 |
| C7 | -0.0001400 | 6230 | GTEx | DepMap | Descartes | 0.02 | 0.47 |
| SCARA5 | -0.0001539 | 6461 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PDGFRA | -0.0001707 | 6706 | GTEx | DepMap | Descartes | 0.23 | 5.67 |
| COL12A1 | -0.0002016 | 7147 | GTEx | DepMap | Descartes | 0.04 | 0.55 |
| PAMR1 | -0.0002086 | 7248 | GTEx | DepMap | Descartes | 0.01 | 0.37 |
| ABCA6 | -0.0002185 | 7394 | GTEx | DepMap | Descartes | 0.01 | 0.24 |
| COL27A1 | -0.0002345 | 7607 | GTEx | DepMap | Descartes | 0.01 | 0.07 |
| ACTA2 | -0.0002612 | 7972 | GTEx | DepMap | Descartes | 0.50 | 36.86 |
Descartes sympathoblasts markers
Top 50 marker genes of sympathoblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/sympathoblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 3.01e-01
Mean rank of genes in gene set: 5969
Median rank of genes in gene set: 6622.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| MGAT4C | 0.0049011 | 27 | GTEx | DepMap | Descartes | 0.90 | 5.63 |
| NTNG1 | 0.0030173 | 83 | GTEx | DepMap | Descartes | 0.76 | 23.89 |
| PCSK1N | 0.0021604 | 160 | GTEx | DepMap | Descartes | 11.96 | 1733.34 |
| GRID2 | 0.0019947 | 184 | GTEx | DepMap | Descartes | 0.34 | 9.14 |
| SPOCK3 | 0.0011203 | 516 | GTEx | DepMap | Descartes | 0.15 | 7.03 |
| SLC35F3 | 0.0007290 | 916 | GTEx | DepMap | Descartes | 0.03 | 1.30 |
| ARC | 0.0006144 | 1124 | GTEx | DepMap | Descartes | 1.95 | 98.35 |
| SORCS3 | 0.0005564 | 1251 | GTEx | DepMap | Descartes | 0.09 | 2.80 |
| CHGB | 0.0005352 | 1306 | GTEx | DepMap | Descartes | 5.71 | 397.12 |
| PCSK2 | 0.0001572 | 2743 | GTEx | DepMap | Descartes | 0.04 | 1.42 |
| GRM7 | 0.0001361 | 2871 | GTEx | DepMap | Descartes | 0.03 | 0.98 |
| KSR2 | 0.0001157 | 3017 | GTEx | DepMap | Descartes | 0.10 | 0.92 |
| CCSER1 | 0.0001022 | 3118 | GTEx | DepMap | Descartes | 0.12 | NA |
| UNC80 | 0.0000311 | 3751 | GTEx | DepMap | Descartes | 0.29 | 3.04 |
| HTATSF1 | 0.0000074 | 4018 | GTEx | DepMap | Descartes | 1.19 | 56.56 |
| PENK | 0.0000062 | 4032 | GTEx | DepMap | Descartes | 0.00 | 0.40 |
| CDH12 | -0.0000186 | 4338 | GTEx | DepMap | Descartes | 0.03 | 1.22 |
| CDH18 | -0.0001252 | 5995 | GTEx | DepMap | Descartes | 0.03 | 0.94 |
| FAM155A | -0.0001419 | 6265 | GTEx | DepMap | Descartes | 0.31 | 4.83 |
| TENM1 | -0.0001907 | 6980 | GTEx | DepMap | Descartes | 0.06 | NA |
| CNTN3 | -0.0001932 | 7029 | GTEx | DepMap | Descartes | 0.00 | 0.13 |
| PACRG | -0.0002136 | 7313 | GTEx | DepMap | Descartes | 0.09 | 7.84 |
| ST18 | -0.0002299 | 7555 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| KCTD16 | -0.0002624 | 7991 | GTEx | DepMap | Descartes | 0.46 | 4.98 |
| ROBO1 | -0.0002713 | 8112 | GTEx | DepMap | Descartes | 0.23 | 4.11 |
| TBX20 | -0.0002753 | 8163 | GTEx | DepMap | Descartes | 0.01 | 1.17 |
| EML6 | -0.0002762 | 8173 | GTEx | DepMap | Descartes | 0.03 | 0.43 |
| SLC24A2 | -0.0003019 | 8504 | GTEx | DepMap | Descartes | 0.00 | 0.06 |
| LAMA3 | -0.0003686 | 9328 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| AGBL4 | -0.0003701 | 9347 | GTEx | DepMap | Descartes | 0.04 | 1.64 |
Descartes erythroblasts markers
Top 50 marker genes of erythroblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/erythroblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 8.64e-01
Mean rank of genes in gene set: 7013.1
Median rank of genes in gene set: 6498
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| SNCA | 0.0005532 | 1261 | GTEx | DepMap | Descartes | 0.67 | 32.02 |
| GCLC | 0.0003281 | 1890 | GTEx | DepMap | Descartes | 0.21 | 8.08 |
| ANK1 | 0.0001389 | 2855 | GTEx | DepMap | Descartes | 0.11 | 1.71 |
| TRAK2 | 0.0001038 | 3111 | GTEx | DepMap | Descartes | 0.17 | 4.05 |
| CPOX | 0.0000908 | 3216 | GTEx | DepMap | Descartes | 0.07 | 3.57 |
| RGS6 | 0.0000305 | 3764 | GTEx | DepMap | Descartes | 0.00 | 0.09 |
| RHD | -0.0000160 | 4299 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SLC25A21 | -0.0000430 | 4683 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SLC4A1 | -0.0000526 | 4836 | GTEx | DepMap | Descartes | 0.00 | 0.07 |
| SPTB | -0.0000626 | 5011 | GTEx | DepMap | Descartes | 0.07 | 1.18 |
| ALAS2 | -0.0000869 | 5372 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TSPAN5 | -0.0001082 | 5703 | GTEx | DepMap | Descartes | 0.75 | 27.06 |
| XPO7 | -0.0001451 | 6328 | GTEx | DepMap | Descartes | 0.22 | 6.03 |
| TMCC2 | -0.0001557 | 6496 | GTEx | DepMap | Descartes | 0.03 | 1.28 |
| ABCB10 | -0.0001558 | 6498 | GTEx | DepMap | Descartes | 0.05 | 1.91 |
| CAT | -0.0001784 | 6810 | GTEx | DepMap | Descartes | 0.31 | 18.56 |
| MICAL2 | -0.0002466 | 7766 | GTEx | DepMap | Descartes | 0.02 | 0.31 |
| SELENBP1 | -0.0002827 | 8243 | GTEx | DepMap | Descartes | 0.01 | 0.36 |
| MARCH3 | -0.0002972 | 8445 | GTEx | DepMap | Descartes | 0.03 | NA |
| FECH | -0.0003285 | 8867 | GTEx | DepMap | Descartes | 0.09 | 1.53 |
| RAPGEF2 | -0.0003700 | 9345 | GTEx | DepMap | Descartes | 0.29 | 5.14 |
| BLVRB | -0.0003703 | 9350 | GTEx | DepMap | Descartes | 0.55 | 41.81 |
| DENND4A | -0.0004055 | 9762 | GTEx | DepMap | Descartes | 0.20 | 3.52 |
| TFR2 | -0.0004099 | 9811 | GTEx | DepMap | Descartes | 0.06 | 2.65 |
| SPECC1 | -0.0006577 | 11479 | GTEx | DepMap | Descartes | 0.03 | 0.36 |
| GYPC | -0.0006717 | 11535 | GTEx | DepMap | Descartes | 0.23 | 15.68 |
| SOX6 | -0.0007868 | 11890 | GTEx | DepMap | Descartes | 0.02 | 0.31 |
| EPB41 | -0.0011033 | 12342 | GTEx | DepMap | Descartes | 0.31 | 6.94 |
| SLC25A37 | -0.0012245 | 12412 | GTEx | DepMap | Descartes | 0.30 | 8.26 |
| NA | NA | NA | NA | NA | NA | NA | NA |
Descartes myeloid markers
Top 50 marker genes of myeloid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/myeloid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 9417.53
Median rank of genes in gene set: 10712.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| FMN1 | 0.0008489 | 757 | GTEx | DepMap | Descartes | 0.35 | 4.15 |
| HRH1 | 0.0002176 | 2383 | GTEx | DepMap | Descartes | 0.05 | 1.65 |
| MSR1 | 0.0001244 | 2954 | GTEx | DepMap | Descartes | 0.26 | 8.94 |
| SLC9A9 | -0.0000278 | 4455 | GTEx | DepMap | Descartes | 0.05 | 1.73 |
| IFNGR1 | -0.0000587 | 4945 | GTEx | DepMap | Descartes | 0.49 | 27.31 |
| WWP1 | -0.0000832 | 5304 | GTEx | DepMap | Descartes | 0.16 | 4.87 |
| PTPRE | -0.0000863 | 5363 | GTEx | DepMap | Descartes | 0.18 | 4.05 |
| RGL1 | -0.0002113 | 7284 | GTEx | DepMap | Descartes | 0.07 | 1.54 |
| SFMBT2 | -0.0002598 | 7945 | GTEx | DepMap | Descartes | 0.04 | 0.68 |
| FGD2 | -0.0002683 | 8073 | GTEx | DepMap | Descartes | 0.02 | 0.31 |
| SLC1A3 | -0.0002992 | 8475 | GTEx | DepMap | Descartes | 0.04 | 1.42 |
| ATP8B4 | -0.0003093 | 8614 | GTEx | DepMap | Descartes | 0.01 | 0.10 |
| MERTK | -0.0003469 | 9090 | GTEx | DepMap | Descartes | 0.06 | 1.50 |
| ABCA1 | -0.0003591 | 9225 | GTEx | DepMap | Descartes | 0.18 | 2.33 |
| MARCH1 | -0.0003821 | 9508 | GTEx | DepMap | Descartes | 0.13 | NA |
| CPVL | -0.0004317 | 10018 | GTEx | DepMap | Descartes | 0.27 | 10.45 |
| SPP1 | -0.0004992 | 10591 | GTEx | DepMap | Descartes | 0.81 | 59.89 |
| HCK | -0.0005047 | 10630 | GTEx | DepMap | Descartes | 0.05 | 2.29 |
| CST3 | -0.0005075 | 10655 | GTEx | DepMap | Descartes | 3.99 | 155.84 |
| LGMN | -0.0005213 | 10770 | GTEx | DepMap | Descartes | 1.03 | 50.06 |
| CD74 | -0.0005800 | 11144 | GTEx | DepMap | Descartes | 2.88 | 93.76 |
| CTSB | -0.0005865 | 11173 | GTEx | DepMap | Descartes | 1.88 | 56.28 |
| AXL | -0.0006405 | 11416 | GTEx | DepMap | Descartes | 0.11 | 2.38 |
| RBPJ | -0.0006452 | 11443 | GTEx | DepMap | Descartes | 1.17 | 28.66 |
| CSF1R | -0.0006560 | 11476 | GTEx | DepMap | Descartes | 0.10 | 2.96 |
| ADAP2 | -0.0006657 | 11514 | GTEx | DepMap | Descartes | 0.13 | 5.18 |
| CD163 | -0.0006852 | 11588 | GTEx | DepMap | Descartes | 0.22 | 4.07 |
| CTSD | -0.0007054 | 11662 | GTEx | DepMap | Descartes | 2.79 | 127.76 |
| SLCO2B1 | -0.0007165 | 11697 | GTEx | DepMap | Descartes | 0.07 | 1.26 |
| ITPR2 | -0.0007169 | 11700 | GTEx | DepMap | Descartes | 0.07 | 0.57 |
Descartes Schwann markers
Top 50 marker genes of Schwann cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/schwann/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 6.38e-01
Mean rank of genes in gene set: 6467.27
Median rank of genes in gene set: 7368
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| NRXN1 | 0.0050922 | 24 | GTEx | DepMap | Descartes | 3.83 | 64.45 |
| SCN7A | 0.0026351 | 108 | GTEx | DepMap | Descartes | 0.69 | 14.39 |
| MARCKS | 0.0025211 | 120 | GTEx | DepMap | Descartes | 15.82 | 546.91 |
| PTN | 0.0019916 | 186 | GTEx | DepMap | Descartes | 1.25 | 124.37 |
| DST | 0.0018315 | 221 | GTEx | DepMap | Descartes | 4.03 | 27.40 |
| SFRP1 | 0.0015089 | 301 | GTEx | DepMap | Descartes | 1.68 | 59.50 |
| PPP2R2B | 0.0010484 | 563 | GTEx | DepMap | Descartes | 0.96 | 13.38 |
| EGFLAM | 0.0007111 | 943 | GTEx | DepMap | Descartes | 0.22 | 7.00 |
| SLC35F1 | 0.0005495 | 1266 | GTEx | DepMap | Descartes | 0.18 | 5.12 |
| IL1RAPL2 | 0.0004152 | 1582 | GTEx | DepMap | Descartes | 0.00 | 0.19 |
| PLCE1 | 0.0002945 | 2022 | GTEx | DepMap | Descartes | 0.14 | 1.91 |
| TRPM3 | 0.0002172 | 2386 | GTEx | DepMap | Descartes | 0.02 | 0.23 |
| IL1RAPL1 | 0.0002030 | 2466 | GTEx | DepMap | Descartes | 0.02 | 0.96 |
| COL25A1 | 0.0001755 | 2628 | GTEx | DepMap | Descartes | 0.13 | 2.22 |
| SORCS1 | 0.0000663 | 3410 | GTEx | DepMap | Descartes | 0.14 | 2.89 |
| PTPRZ1 | 0.0000039 | 4061 | GTEx | DepMap | Descartes | 0.01 | 0.34 |
| HMGA2 | -0.0000113 | 4242 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MDGA2 | -0.0000179 | 4330 | GTEx | DepMap | Descartes | 0.00 | 0.06 |
| NRXN3 | -0.0000492 | 4775 | GTEx | DepMap | Descartes | 0.03 | 0.45 |
| STARD13 | -0.0000810 | 5271 | GTEx | DepMap | Descartes | 0.05 | 0.89 |
| LRRTM4 | -0.0001714 | 6714 | GTEx | DepMap | Descartes | 0.03 | 1.42 |
| PLP1 | -0.0002051 | 7194 | GTEx | DepMap | Descartes | 0.03 | 2.30 |
| ERBB3 | -0.0002292 | 7542 | GTEx | DepMap | Descartes | 0.01 | 0.37 |
| XKR4 | -0.0002355 | 7618 | GTEx | DepMap | Descartes | 0.02 | 0.19 |
| COL5A2 | -0.0002406 | 7685 | GTEx | DepMap | Descartes | 0.13 | 2.52 |
| MPZ | -0.0002588 | 7928 | GTEx | DepMap | Descartes | 0.03 | 2.79 |
| SOX5 | -0.0003243 | 8808 | GTEx | DepMap | Descartes | 0.36 | 7.55 |
| ADAMTS5 | -0.0003720 | 9378 | GTEx | DepMap | Descartes | 0.01 | 0.09 |
| ERBB4 | -0.0003900 | 9596 | GTEx | DepMap | Descartes | 0.02 | 0.26 |
| GAS7 | -0.0004381 | 10077 | GTEx | DepMap | Descartes | 0.06 | 1.29 |
Descartes Megakaryocytes markers
Top 50 marker genes of Megakaryocytes cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/megakaryocytes/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.17e-01
Mean rank of genes in gene set: 7023.27
Median rank of genes in gene set: 7131
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ANGPT1 | 0.0018779 | 205 | GTEx | DepMap | Descartes | 0.16 | 5.42 |
| CD9 | 0.0016792 | 253 | GTEx | DepMap | Descartes | 2.04 | 176.59 |
| PDE3A | 0.0013906 | 357 | GTEx | DepMap | Descartes | 0.25 | 4.87 |
| MCTP1 | 0.0006794 | 1004 | GTEx | DepMap | Descartes | 0.06 | 1.46 |
| HIPK2 | 0.0006615 | 1039 | GTEx | DepMap | Descartes | 0.75 | 6.83 |
| PRKAR2B | 0.0005535 | 1259 | GTEx | DepMap | Descartes | 0.88 | 36.31 |
| LIMS1 | 0.0000994 | 3137 | GTEx | DepMap | Descartes | 1.01 | 30.50 |
| DOK6 | 0.0000803 | 3298 | GTEx | DepMap | Descartes | 0.17 | 2.57 |
| LTBP1 | 0.0000280 | 3785 | GTEx | DepMap | Descartes | 0.04 | 0.70 |
| INPP4B | 0.0000233 | 3827 | GTEx | DepMap | Descartes | 0.10 | 1.63 |
| ITGB3 | 0.0000221 | 3839 | GTEx | DepMap | Descartes | 0.00 | 0.02 |
| SLC24A3 | -0.0000075 | 4188 | GTEx | DepMap | Descartes | 0.01 | 0.23 |
| ITGA2B | -0.0000309 | 4509 | GTEx | DepMap | Descartes | 0.01 | 0.46 |
| MED12L | -0.0000774 | 5216 | GTEx | DepMap | Descartes | 0.04 | 0.56 |
| P2RX1 | -0.0001067 | 5675 | GTEx | DepMap | Descartes | 0.00 | 0.02 |
| ACTB | -0.0001230 | 5963 | GTEx | DepMap | Descartes | 32.24 | 2011.59 |
| TRPC6 | -0.0001281 | 6040 | GTEx | DepMap | Descartes | 0.01 | 0.22 |
| TUBB1 | -0.0001352 | 6156 | GTEx | DepMap | Descartes | 0.00 | 0.09 |
| ARHGAP6 | -0.0001412 | 6252 | GTEx | DepMap | Descartes | 0.01 | 0.12 |
| GP1BA | -0.0001448 | 6323 | GTEx | DepMap | Descartes | 0.00 | 0.02 |
| STON2 | -0.0001789 | 6817 | GTEx | DepMap | Descartes | 0.08 | 2.68 |
| UBASH3B | -0.0001908 | 6984 | GTEx | DepMap | Descartes | 0.04 | 0.84 |
| ACTN1 | -0.0002005 | 7131 | GTEx | DepMap | Descartes | 0.74 | 23.02 |
| VCL | -0.0002118 | 7291 | GTEx | DepMap | Descartes | 0.20 | 3.33 |
| RAP1B | -0.0002201 | 7412 | GTEx | DepMap | Descartes | 1.05 | 11.12 |
| PSTPIP2 | -0.0002514 | 7827 | GTEx | DepMap | Descartes | 0.03 | 0.86 |
| TLN1 | -0.0002928 | 8378 | GTEx | DepMap | Descartes | 0.45 | 6.62 |
| MMRN1 | -0.0002988 | 8466 | GTEx | DepMap | Descartes | 0.01 | 0.31 |
| RAB27B | -0.0003253 | 8826 | GTEx | DepMap | Descartes | 0.02 | 0.31 |
| SPN | -0.0003277 | 8855 | GTEx | DepMap | Descartes | 0.01 | 0.20 |
Descartes Lyphoid markers
Top 50 marker genes of Lyphoid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/lymphoid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.73e-01
Mean rank of genes in gene set: 7350.71
Median rank of genes in gene set: 9172
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TMSB10 | 0.0030627 | 79 | GTEx | DepMap | Descartes | 102.83 | 32472.88 |
| STK39 | 0.0013825 | 361 | GTEx | DepMap | Descartes | 0.79 | 36.81 |
| ARID5B | 0.0009763 | 639 | GTEx | DepMap | Descartes | 0.67 | 13.98 |
| BCL2 | 0.0009714 | 645 | GTEx | DepMap | Descartes | 0.80 | 16.62 |
| BACH2 | 0.0004853 | 1419 | GTEx | DepMap | Descartes | 0.32 | 5.63 |
| PITPNC1 | 0.0004667 | 1464 | GTEx | DepMap | Descartes | 0.38 | 8.74 |
| SP100 | 0.0003587 | 1761 | GTEx | DepMap | Descartes | 0.26 | 6.73 |
| ABLIM1 | 0.0003061 | 1979 | GTEx | DepMap | Descartes | 0.43 | 7.69 |
| GNG2 | 0.0001587 | 2732 | GTEx | DepMap | Descartes | 0.88 | 35.94 |
| SORL1 | 0.0001284 | 2932 | GTEx | DepMap | Descartes | 0.36 | 4.99 |
| FYN | 0.0000660 | 3414 | GTEx | DepMap | Descartes | 1.58 | 66.91 |
| MCTP2 | 0.0000517 | 3548 | GTEx | DepMap | Descartes | 0.07 | 1.23 |
| B2M | 0.0000365 | 3691 | GTEx | DepMap | Descartes | 33.79 | 1879.39 |
| CCND3 | 0.0000058 | 4036 | GTEx | DepMap | Descartes | 0.31 | 18.73 |
| EVL | -0.0000338 | 4554 | GTEx | DepMap | Descartes | 2.54 | 98.60 |
| NCALD | -0.0000389 | 4632 | GTEx | DepMap | Descartes | 0.12 | 4.59 |
| ITPKB | -0.0002456 | 7753 | GTEx | DepMap | Descartes | 0.01 | 0.13 |
| SAMD3 | -0.0002465 | 7765 | GTEx | DepMap | Descartes | 0.01 | 0.35 |
| ANKRD44 | -0.0002634 | 8004 | GTEx | DepMap | Descartes | 0.16 | 3.36 |
| WIPF1 | -0.0003321 | 8915 | GTEx | DepMap | Descartes | 0.19 | 5.62 |
| PLEKHA2 | -0.0003475 | 9099 | GTEx | DepMap | Descartes | 0.04 | 0.88 |
| SKAP1 | -0.0003604 | 9245 | GTEx | DepMap | Descartes | 0.01 | 0.75 |
| PRKCH | -0.0003718 | 9373 | GTEx | DepMap | Descartes | 0.01 | 0.44 |
| CCL5 | -0.0003734 | 9395 | GTEx | DepMap | Descartes | 0.12 | 15.32 |
| SCML4 | -0.0003803 | 9481 | GTEx | DepMap | Descartes | 0.07 | 2.33 |
| LEF1 | -0.0003881 | 9573 | GTEx | DepMap | Descartes | 0.02 | 1.07 |
| RCSD1 | -0.0004325 | 10033 | GTEx | DepMap | Descartes | 0.02 | 0.41 |
| IKZF1 | -0.0004489 | 10181 | GTEx | DepMap | Descartes | 0.03 | 0.47 |
| ETS1 | -0.0004532 | 10221 | GTEx | DepMap | Descartes | 0.06 | 1.46 |
| ARHGAP15 | -0.0005213 | 10768 | GTEx | DepMap | Descartes | 0.04 | 1.30 |
Below shows the ranks on this GEP for immune marker gene lists curated by CellTypist (https://www.celltypist.org/encyclopedia).
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Epithelial cells: Epithelial cells (model markers)
highly specialised cells which are located in the outer layer of stroma and form the cellular components of the epithelium:
Wilcoxon ranksum test P-value for gene set overrepresentation: 5.74e-02
Mean rank of genes in gene set: 2235
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| WFDC2 | 0.0006825 | 998 | GTEx | DepMap | Descartes | 0.27 | 52.20 |
| CDH3 | 0.0000599 | 3472 | GTEx | DepMap | Descartes | 0.01 | 0.61 |
Megakaryocyte precursor: Megakaryocyte precursor (model markers)
megakaryocyte precursors in the bone marrow that are committed to megakaryocytes:
Wilcoxon ranksum test P-value for gene set overrepresentation: 6.27e-02
Mean rank of genes in gene set: 2349.5
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ADRA2A | 0.0026996 | 101 | GTEx | DepMap | Descartes | 0.07 | 2.53 |
| SLC10A5 | -0.0000365 | 4598 | GTEx | DepMap | Descartes | 0.01 | 0.35 |
Mast cells: Mast cells (model markers)
long-lived innate migrant cells found in most tissues with many large basophilic granules rich in histamine and heparin:
Wilcoxon ranksum test P-value for gene set overrepresentation: 6.46e-02
Mean rank of genes in gene set: 777
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CPA3 | 0.0008332 | 777 | GTEx | DepMap | Descartes | 0 | 0.05 |