Program: 12. Cell Cycle.
Submit a comment on this gene expression program’s interpretation: CLICK
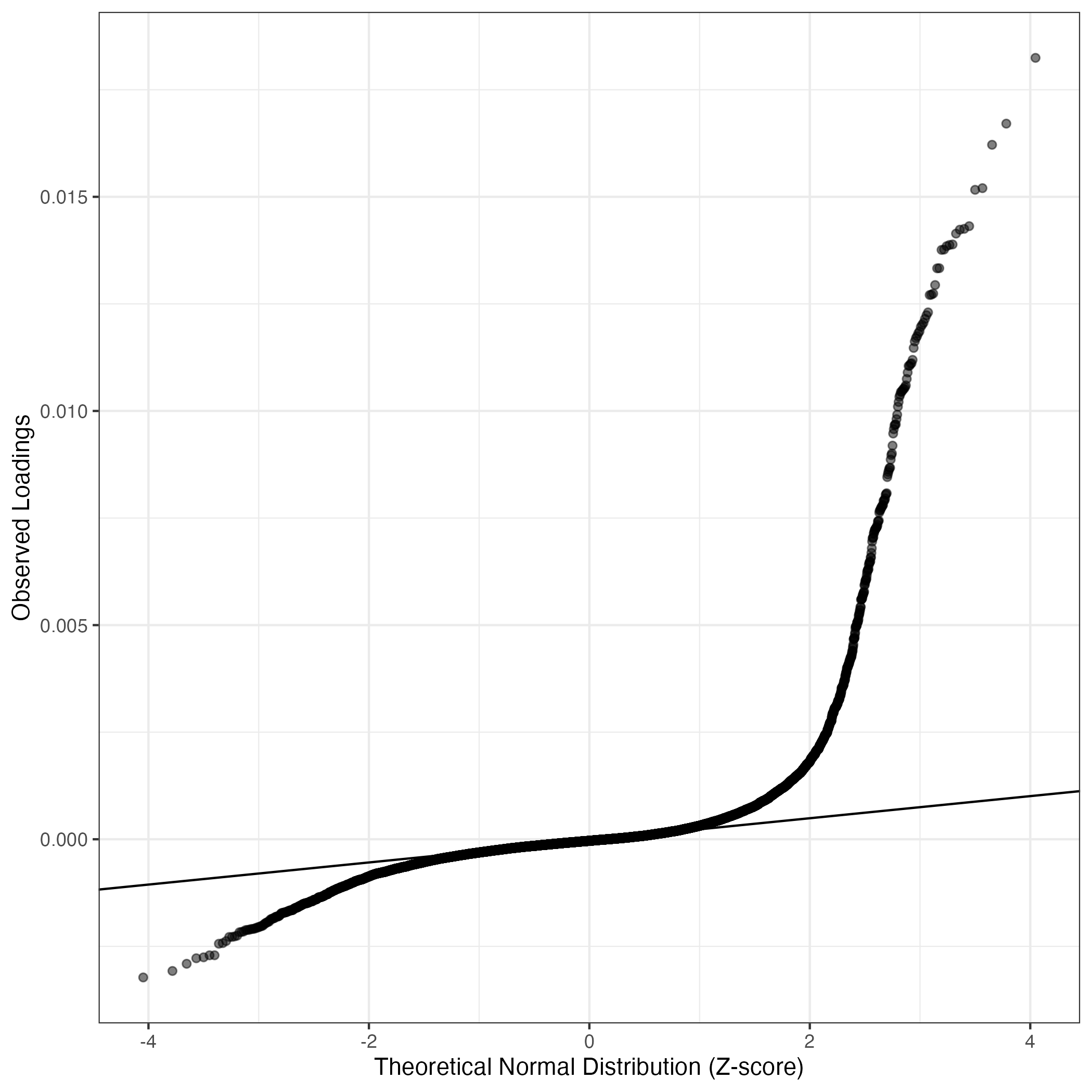
QQ-plot of gene loadings, averaged over both independent splits of the data
This plot highlights the relative contribution of each gene to the GEP
Top genes driving this program.
Note: Decartes website is buggy, try refreshing. Also, Decartes fetal adrenal data have been collected at specific time points (89-122 days), all possible cell types of interest may not be represented, do not overinterpret.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
| Gene | Loading | Gene.Name | GTEx | DepMap | Descartes | Mean.Counts | Mean.Tpm | |
|---|---|---|---|---|---|---|---|---|
| 1 | UBE2C | 0.0182446 | ubiquitin conjugating enzyme E2 C | GTEx | DepMap | Descartes | 26.85 | 473.51 |
| 2 | CKS2 | 0.0167088 | CDC28 protein kinase regulatory subunit 2 | GTEx | DepMap | Descartes | 13.39 | 340.97 |
| 3 | CENPF | 0.0162148 | centromere protein F | GTEx | DepMap | Descartes | 18.30 | 61.44 |
| 4 | CDK1 | 0.0152023 | cyclin dependent kinase 1 | GTEx | DepMap | Descartes | 9.49 | 71.41 |
| 5 | HMGB2 | 0.0151639 | high mobility group box 2 | GTEx | DepMap | Descartes | 48.61 | 1588.50 |
| 6 | CDC20 | 0.0143149 | cell division cycle 20 | GTEx | DepMap | Descartes | 8.05 | 288.27 |
| 7 | CENPE | 0.0142521 | centromere protein E | GTEx | DepMap | Descartes | 8.27 | 22.42 |
| 8 | TPX2 | 0.0142331 | TPX2 microtubule nucleation factor | GTEx | DepMap | Descartes | 8.23 | 28.23 |
| 9 | CCNB1 | 0.0141412 | cyclin B1 | GTEx | DepMap | Descartes | 8.58 | 193.34 |
| 10 | H2AFX | 0.0138880 | NA | GTEx | DepMap | Descartes | 23.36 | 2577.17 |
| 11 | BIRC5 | 0.0138758 | baculoviral IAP repeat containing 5 | GTEx | DepMap | Descartes | 13.71 | 338.97 |
| 12 | TOP2A | 0.0138475 | DNA topoisomerase II alpha | GTEx | DepMap | Descartes | 25.38 | 128.60 |
| 13 | NUSAP1 | 0.0137677 | nucleolar and spindle associated protein 1 | GTEx | DepMap | Descartes | 6.87 | 35.45 |
| 14 | PRC1 | 0.0137612 | protein regulator of cytokinesis 1 | GTEx | DepMap | Descartes | 7.82 | 57.16 |
| 15 | CENPA | 0.0133333 | centromere protein A | GTEx | DepMap | Descartes | 12.06 | 228.50 |
| 16 | MKI67 | 0.0133305 | marker of proliferation Ki-67 | GTEx | DepMap | Descartes | 13.00 | 72.38 |
| 17 | CCNA2 | 0.0129406 | cyclin A2 | GTEx | DepMap | Descartes | 7.21 | 161.02 |
| 18 | KIF23 | 0.0127404 | kinesin family member 23 | GTEx | DepMap | Descartes | 3.37 | 16.72 |
| 19 | PLK1 | 0.0127155 | polo like kinase 1 | GTEx | DepMap | Descartes | 4.41 | 69.56 |
| 20 | HMMR | 0.0127081 | hyaluronan mediated motility receptor | GTEx | DepMap | Descartes | 4.79 | 24.39 |
| 21 | CKAP2L | 0.0123016 | cytoskeleton associated protein 2 like | GTEx | DepMap | Descartes | 4.38 | 23.85 |
| 22 | AURKA | 0.0122353 | aurora kinase A | GTEx | DepMap | Descartes | 2.98 | 31.90 |
| 23 | KIF11 | 0.0121501 | kinesin family member 11 | GTEx | DepMap | Descartes | 4.31 | 14.21 |
| 24 | CDCA8 | 0.0120593 | cell division cycle associated 8 | GTEx | DepMap | Descartes | 8.51 | 63.88 |
| 25 | SMC4 | 0.0120123 | structural maintenance of chromosomes 4 | GTEx | DepMap | Descartes | 10.17 | 48.44 |
| 26 | TACC3 | 0.0119629 | transforming acidic coiled-coil containing protein 3 | GTEx | DepMap | Descartes | 5.13 | 38.48 |
| 27 | KPNA2 | 0.0118593 | karyopherin subunit alpha 2 | GTEx | DepMap | Descartes | 3.06 | 42.20 |
| 28 | BUB1B | 0.0118147 | BUB1 mitotic checkpoint serine/threonine kinase B | GTEx | DepMap | Descartes | 3.40 | 11.56 |
| 29 | ASPM | 0.0117468 | assembly factor for spindle microtubules | GTEx | DepMap | Descartes | 4.21 | 17.60 |
| 30 | AURKB | 0.0116996 | aurora kinase B | GTEx | DepMap | Descartes | 3.84 | 88.61 |
| 31 | KIF20A | 0.0116224 | kinesin family member 20A | GTEx | DepMap | Descartes | 3.69 | 69.34 |
| 32 | SGOL2A | 0.0114722 | NA | GTEx | DepMap | Descartes | 3.13 | NA |
| 33 | MIS18BP1 | 0.0111932 | MIS18 binding protein 1 | GTEx | DepMap | Descartes | 3.66 | 14.66 |
| 34 | TUBB4B | 0.0111140 | tubulin beta 4B class IVb | GTEx | DepMap | Descartes | 23.69 | 1585.68 |
| 35 | UBE2S | 0.0110995 | ubiquitin conjugating enzyme E2 S | GTEx | DepMap | Descartes | 21.27 | 171.16 |
| 36 | CDCA3 | 0.0110560 | cell division cycle associated 3 | GTEx | DepMap | Descartes | 5.07 | 184.58 |
| 37 | ARL6IP1 | 0.0110511 | ADP ribosylation factor like GTPase 6 interacting protein 1 | GTEx | DepMap | Descartes | 24.32 | 376.73 |
| 38 | KIF22 | 0.0108983 | kinesin family member 22 | GTEx | DepMap | Descartes | 3.23 | 36.05 |
| 39 | INCENP | 0.0107483 | inner centromere protein | GTEx | DepMap | Descartes | 4.99 | 30.23 |
| 40 | CASC5 | 0.0105975 | NA | GTEx | DepMap | Descartes | 3.86 | NA |
| 41 | PTTG1 | 0.0105564 | PTTG1 regulator of sister chromatid separation, securin | GTEx | DepMap | Descartes | 6.17 | 165.22 |
| 42 | TUBA1C | 0.0105203 | tubulin alpha 1c | GTEx | DepMap | Descartes | 6.69 | 157.12 |
| 43 | DLGAP5 | 0.0105125 | DLG associated protein 5 | GTEx | DepMap | Descartes | 2.83 | 14.01 |
| 44 | FAM64A | 0.0104793 | NA | GTEx | DepMap | Descartes | 2.88 | NA |
| 45 | KIF2C | 0.0104493 | kinesin family member 2C | GTEx | DepMap | Descartes | 2.53 | 17.71 |
| 46 | TUBA1B | 0.0104459 | tubulin alpha 1b | GTEx | DepMap | Descartes | 58.85 | 2696.16 |
| 47 | KIF20B | 0.0103793 | kinesin family member 20B | GTEx | DepMap | Descartes | 3.38 | 9.63 |
| 48 | CCNB2 | 0.0103295 | cyclin B2 | GTEx | DepMap | Descartes | 5.23 | 59.15 |
| 49 | SMC2 | 0.0102096 | structural maintenance of chromosomes 2 | GTEx | DepMap | Descartes | 9.42 | 28.60 |
| 50 | SPC25 | 0.0101009 | SPC25 component of NDC80 kinetochore complex | GTEx | DepMap | Descartes | 4.52 | 61.60 |
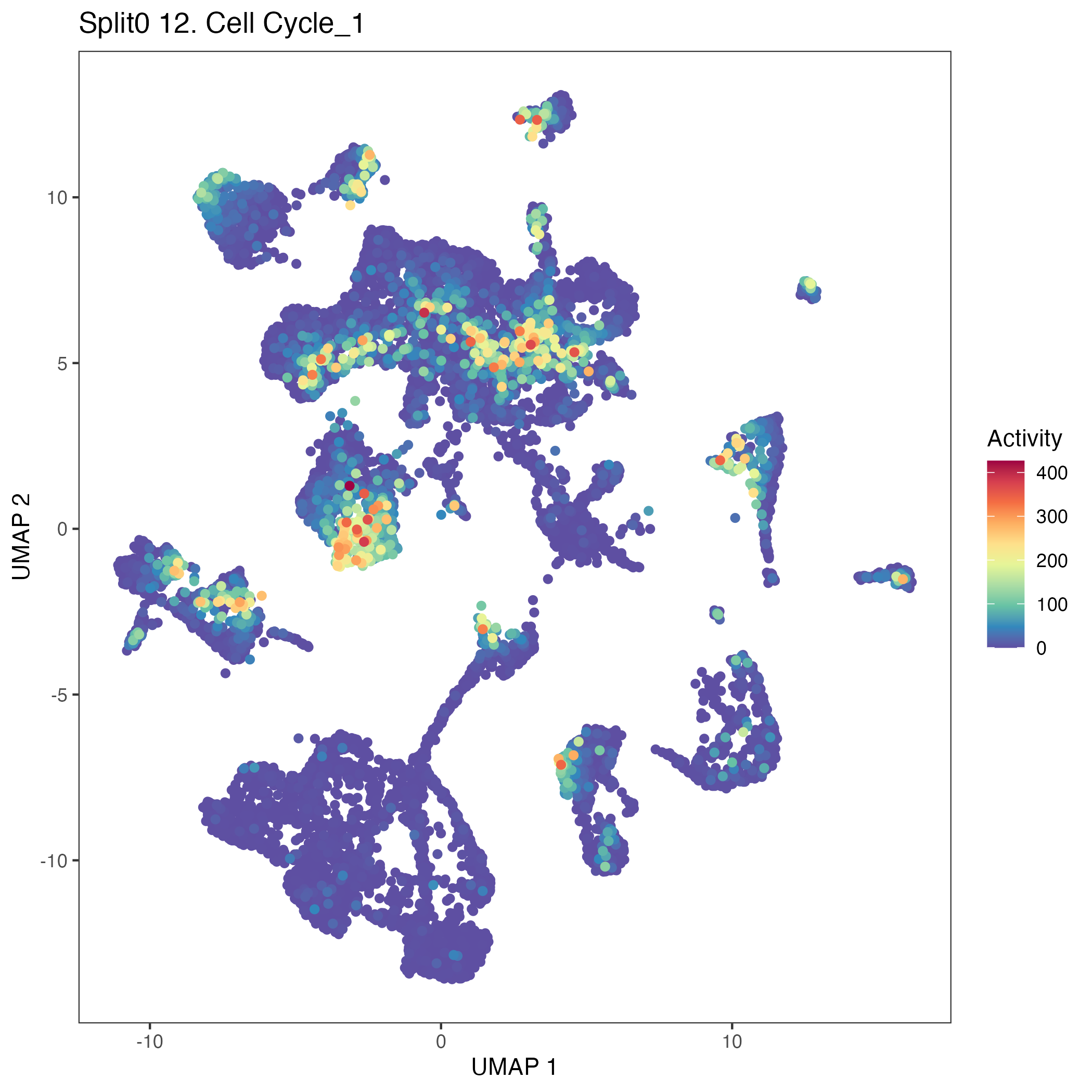
UMAP plots showing activity of gene expression program identified in GEP 12. Cell Cycle:
Gene set Enrichments for this program, caculated from top 50 genes
mSigDB Cell Types Gene Set:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| FAN_EMBRYONIC_CTX_MICROGLIA_1 | 6.21e-80 | 772.89 | 355.99 | 1.39e-77 | 4.16e-77 | 41UBE2C, CKS2, CENPF, CDK1, HMGB2, CDC20, CENPE, TPX2, CCNB1, BIRC5, TOP2A, NUSAP1, PRC1, MKI67, CCNA2, KIF23, PLK1, HMMR, CKAP2L, AURKA, KIF11, CDCA8, SMC4, TACC3, KPNA2, BUB1B, ASPM, AURKB, KIF20A, TUBB4B, CDCA3, ARL6IP1, KIF22, PTTG1, TUBA1C, DLGAP5, KIF2C, KIF20B, CCNB2, SMC2, SPC25 |
155 |
| ZHONG_PFC_C1_OPC | 1.67e-82 | 898.28 | 350.53 | 1.12e-79 | 1.12e-79 | 45UBE2C, CKS2, CENPF, CDK1, HMGB2, CDC20, CENPE, TPX2, CCNB1, BIRC5, TOP2A, NUSAP1, PRC1, CENPA, MKI67, CCNA2, KIF23, PLK1, HMMR, CKAP2L, AURKA, KIF11, CDCA8, SMC4, TACC3, KPNA2, BUB1B, ASPM, AURKB, KIF20A, MIS18BP1, TUBB4B, UBE2S, CDCA3, ARL6IP1, KIF22, PTTG1, TUBA1C, DLGAP5, KIF2C, TUBA1B, KIF20B, CCNB2, SMC2, SPC25 |
238 |
| ZHONG_PFC_C2_UNKNOWN_NPC | 3.63e-62 | 621.17 | 315.56 | 6.10e-60 | 2.44e-59 | 30UBE2C, CKS2, CENPF, CDC20, CENPE, TPX2, CCNB1, TOP2A, NUSAP1, MKI67, CCNA2, PLK1, CKAP2L, AURKA, CDCA8, TACC3, KPNA2, ASPM, AURKB, KIF20A, MIS18BP1, TUBB4B, CDCA3, ARL6IP1, KIF22, PTTG1, TUBA1C, DLGAP5, KIF2C, CCNB2 |
76 |
| FAN_EMBRYONIC_CTX_NSC_2 | 4.51e-80 | 759.42 | 309.82 | 1.39e-77 | 3.03e-77 | 44UBE2C, CKS2, CENPF, CDK1, HMGB2, CDC20, CENPE, TPX2, CCNB1, BIRC5, TOP2A, NUSAP1, PRC1, CENPA, MKI67, CCNA2, KIF23, PLK1, HMMR, CKAP2L, AURKA, KIF11, CDCA8, SMC4, TACC3, KPNA2, BUB1B, ASPM, AURKB, KIF20A, MIS18BP1, TUBB4B, UBE2S, CDCA3, ARL6IP1, KIF22, PTTG1, TUBA1C, DLGAP5, KIF2C, KIF20B, CCNB2, SMC2, SPC25 |
233 |
| ZHONG_PFC_C8_ORG_PROLIFERATING | 3.89e-46 | 416.83 | 209.08 | 2.37e-44 | 2.61e-43 | 23CKS2, CENPF, CDC20, CENPE, TPX2, CCNB1, BIRC5, NUSAP1, PRC1, CENPA, PLK1, AURKA, CDCA8, TACC3, BUB1B, ASPM, KIF20A, TUBB4B, CDCA3, ARL6IP1, PTTG1, TUBA1C, CCNB2 |
62 |
| ZHONG_PFC_MAJOR_TYPES_NPCS | 4.85e-60 | 341.20 | 178.92 | 6.51e-58 | 3.26e-57 | 33UBE2C, CKS2, CENPF, CDK1, HMGB2, CDC20, TPX2, CCNB1, BIRC5, TOP2A, NUSAP1, PRC1, MKI67, CCNA2, KIF23, CKAP2L, KIF11, SMC4, TACC3, KPNA2, ASPM, AURKB, MIS18BP1, TUBB4B, CDCA3, KIF22, PTTG1, DLGAP5, KIF2C, TUBA1B, CCNB2, SMC2, SPC25 |
142 |
| ZHONG_PFC_C3_UNKNOWN_INP | 2.18e-30 | 354.58 | 159.81 | 9.13e-29 | 1.46e-27 | 15UBE2C, CENPF, CDK1, CENPE, TPX2, BIRC5, PRC1, CCNA2, KIF23, CKAP2L, CDCA8, KPNA2, DLGAP5, CCNB2, SPC25 |
38 |
| MANNO_MIDBRAIN_NEUROTYPES_HPROGFPM | 1.42e-54 | 171.80 | 88.09 | 1.59e-52 | 9.56e-52 | 37UBE2C, CKS2, CENPF, CDK1, HMGB2, CENPE, TPX2, CCNB1, BIRC5, TOP2A, NUSAP1, PRC1, MKI67, CCNA2, KIF23, PLK1, HMMR, CKAP2L, AURKA, KIF11, CDCA8, SMC4, TACC3, KPNA2, BUB1B, ASPM, AURKB, MIS18BP1, UBE2S, CDCA3, KIF22, PTTG1, DLGAP5, KIF2C, CCNB2, SMC2, SPC25 |
356 |
| TRAVAGLINI_LUNG_PROLIFERATING_NK_T_CELL | 9.25e-38 | 154.27 | 81.62 | 4.77e-36 | 6.20e-35 | 23UBE2C, CKS2, CENPF, CDK1, HMGB2, CDC20, TPX2, BIRC5, TOP2A, MKI67, CCNA2, HMMR, CDCA8, SMC4, TACC3, ASPM, AURKB, TUBB4B, CDCA3, KIF22, KIF2C, TUBA1B, SMC2 |
129 |
| MANNO_MIDBRAIN_NEUROTYPES_HPROGBP | 1.08e-50 | 153.46 | 81.33 | 9.03e-49 | 7.22e-48 | 34UBE2C, CKS2, CENPF, CDK1, HMGB2, CENPE, TPX2, CCNB1, BIRC5, TOP2A, NUSAP1, PRC1, MKI67, CCNA2, KIF23, PLK1, HMMR, CKAP2L, AURKA, KIF11, CDCA8, SMC4, TACC3, BUB1B, ASPM, AURKB, MIS18BP1, CDCA3, KIF22, PTTG1, DLGAP5, KIF2C, CCNB2, SPC25 |
300 |
| MANNO_MIDBRAIN_NEUROTYPES_HPROGFPL | 1.44e-49 | 141.22 | 75.17 | 1.08e-47 | 9.69e-47 | 34UBE2C, CENPF, CDK1, HMGB2, CENPE, TPX2, CCNB1, BIRC5, TOP2A, NUSAP1, PRC1, MKI67, CCNA2, KIF23, PLK1, HMMR, CKAP2L, AURKA, KIF11, CDCA8, SMC4, TACC3, BUB1B, ASPM, AURKB, MIS18BP1, CDCA3, KIF22, PTTG1, DLGAP5, KIF2C, CCNB2, SMC2, SPC25 |
323 |
| ZHONG_PFC_C1_MICROGLIA | 2.29e-44 | 126.87 | 68.79 | 1.28e-42 | 1.54e-41 | 30UBE2C, CKS2, CENPF, CDK1, HMGB2, TPX2, CCNB1, BIRC5, TOP2A, NUSAP1, PRC1, CCNA2, KIF23, PLK1, HMMR, CKAP2L, AURKA, KIF11, CDCA8, TACC3, KPNA2, ASPM, AURKB, CDCA3, KIF22, KIF2C, TUBA1B, KIF20B, CCNB2, SPC25 |
257 |
| GAO_LARGE_INTESTINE_24W_C2_MKI67POS_PROGENITOR | 1.78e-30 | 125.26 | 64.48 | 7.97e-29 | 1.20e-27 | 19UBE2C, CENPF, CDK1, CDC20, TPX2, CCNB1, BIRC5, TOP2A, NUSAP1, PRC1, MKI67, CCNA2, TACC3, ASPM, AURKB, PTTG1, DLGAP5, KIF20B, CCNB2 |
113 |
| TRAVAGLINI_LUNG_PROLIFERATING_BASAL_CELL | 1.38e-51 | 139.89 | 62.14 | 1.32e-49 | 9.27e-49 | 43UBE2C, CKS2, CENPF, CDK1, HMGB2, CDC20, CENPE, TPX2, CCNB1, BIRC5, TOP2A, NUSAP1, PRC1, CENPA, MKI67, CCNA2, KIF23, PLK1, HMMR, CKAP2L, AURKA, KIF11, SMC4, TACC3, KPNA2, BUB1B, ASPM, AURKB, KIF20A, MIS18BP1, UBE2S, CDCA3, ARL6IP1, KIF22, PTTG1, TUBA1C, DLGAP5, KIF2C, TUBA1B, KIF20B, CCNB2, SMC2, SPC25 |
891 |
| GAO_LARGE_INTESTINE_ADULT_CH_MKI67HIGH_CELLS | 7.51e-29 | 121.39 | 61.86 | 2.80e-27 | 5.04e-26 | 18CENPF, TPX2, BIRC5, TOP2A, CENPA, MKI67, CCNA2, KIF23, HMMR, CKAP2L, KIF11, CDCA8, TACC3, BUB1B, AURKB, KIF20A, CDCA3, KIF2C |
107 |
| TRAVAGLINI_LUNG_PROLIFERATING_MACROPHAGE_CELL | 4.92e-48 | 109.55 | 50.51 | 3.30e-46 | 3.30e-45 | 42UBE2C, CKS2, CENPF, CDK1, HMGB2, CDC20, CENPE, TPX2, CCNB1, BIRC5, TOP2A, NUSAP1, PRC1, CENPA, MKI67, CCNA2, KIF23, PLK1, HMMR, CKAP2L, AURKA, KIF11, SMC4, TACC3, KPNA2, ASPM, AURKB, MIS18BP1, TUBB4B, UBE2S, CDCA3, ARL6IP1, KIF22, PTTG1, TUBA1C, DLGAP5, KIF2C, TUBA1B, KIF20B, CCNB2, SMC2, SPC25 |
968 |
| MANNO_MIDBRAIN_NEUROTYPES_HNPROG | 9.99e-32 | 79.79 | 42.87 | 4.79e-30 | 6.70e-29 | 23UBE2C, CENPF, CDK1, HMGB2, TPX2, BIRC5, TOP2A, NUSAP1, PRC1, MKI67, CCNA2, KIF23, PLK1, CKAP2L, KIF11, CDCA8, TACC3, BUB1B, ASPM, CDCA3, DLGAP5, KIF2C, CCNB2 |
229 |
| BUSSLINGER_ESOPHAGEAL_EARLY_SUPRABASAL_CELLS | 8.10e-17 | 79.76 | 35.34 | 2.59e-15 | 5.43e-14 | 11CENPF, CDK1, HMGB2, CCNB1, TOP2A, PRC1, MKI67, SMC4, ASPM, PTTG1, TUBA1B |
79 |
| BUSSLINGER_ESOPHAGEAL_PROLIFERATING_BASAL_CELLS | 1.75e-15 | 80.39 | 34.14 | 5.35e-14 | 1.18e-12 | 10CENPF, HMGB2, TOP2A, NUSAP1, PRC1, MKI67, SMC4, ASPM, ARL6IP1, TUBA1B |
70 |
| HAY_BONE_MARROW_PRO_B | 7.48e-29 | 58.34 | 31.58 | 2.80e-27 | 5.02e-26 | 23UBE2C, CENPF, HMGB2, TPX2, BIRC5, TOP2A, NUSAP1, CENPA, MKI67, CKAP2L, KIF11, CDCA8, SMC4, TACC3, KPNA2, ASPM, AURKB, MIS18BP1, UBE2S, ARL6IP1, INCENP, TUBA1B, SPC25 |
304 |
Dowload full table
mSigDB Hallmark Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| HALLMARK_G2M_CHECKPOINT | 4.78e-50 | 186.50 | 99.16 | 2.39e-48 | 2.39e-48 | 31UBE2C, CKS2, CENPF, CDK1, CDC20, CENPE, TPX2, BIRC5, TOP2A, NUSAP1, PRC1, CENPA, MKI67, CCNA2, KIF23, PLK1, HMMR, AURKA, KIF11, SMC4, TACC3, KPNA2, AURKB, UBE2S, KIF22, INCENP, PTTG1, KIF2C, KIF20B, CCNB2, SMC2 |
200 |
| HALLMARK_E2F_TARGETS | 3.92e-37 | 109.67 | 59.31 | 9.80e-36 | 1.96e-35 | 25CKS2, CDK1, HMGB2, CDC20, CENPE, BIRC5, TOP2A, MKI67, PLK1, HMMR, AURKA, CDCA8, SMC4, TACC3, KPNA2, BUB1B, AURKB, UBE2S, CDCA3, KIF22, PTTG1, DLGAP5, KIF2C, CCNB2, SPC25 |
200 |
| HALLMARK_MITOTIC_SPINDLE | 1.38e-25 | 65.66 | 34.33 | 2.29e-24 | 6.88e-24 | 19CENPF, CDK1, CENPE, TPX2, BIRC5, TOP2A, NUSAP1, PRC1, KIF23, PLK1, AURKA, KIF11, SMC4, KIF22, INCENP, DLGAP5, KIF2C, KIF20B, CCNB2 |
199 |
| HALLMARK_GLYCOLYSIS | 1.56e-04 | 10.98 | 3.37 | 1.95e-03 | 7.81e-03 | 5CDK1, CENPA, HMMR, AURKA, KIF20A |
200 |
| HALLMARK_SPERMATOGENESIS | 3.98e-04 | 12.79 | 3.30 | 3.98e-03 | 1.99e-02 | 4CDK1, AURKA, KIF2C, CCNB2 |
135 |
| HALLMARK_ESTROGEN_RESPONSE_LATE | 1.46e-02 | 6.25 | 1.23 | 1.04e-01 | 7.29e-01 | 3CDC20, TOP2A, KIF20A |
200 |
| HALLMARK_MYC_TARGETS_V1 | 1.46e-02 | 6.25 | 1.23 | 1.04e-01 | 7.29e-01 | 3CDC20, CCNA2, KPNA2 |
200 |
| HALLMARK_APOPTOSIS | 6.42e-02 | 5.05 | 0.59 | 4.01e-01 | 1.00e+00 | 2HMGB2, TOP2A |
161 |
| HALLMARK_MTORC1_SIGNALING | 9.29e-02 | 4.06 | 0.47 | 5.16e-01 | 1.00e+00 | 2PLK1, AURKA |
200 |
| HALLMARK_MYC_TARGETS_V2 | 1.39e-01 | 6.90 | 0.17 | 6.96e-01 | 1.00e+00 | 1PLK1 |
58 |
| HALLMARK_PEROXISOME | 2.36e-01 | 3.82 | 0.09 | 9.90e-01 | 1.00e+00 | 1TOP2A |
104 |
| HALLMARK_PI3K_AKT_MTOR_SIGNALING | 2.38e-01 | 3.78 | 0.09 | 9.90e-01 | 1.00e+00 | 1CDK1 |
105 |
| HALLMARK_DNA_REPAIR | 3.21e-01 | 2.64 | 0.07 | 1.00e+00 | 1.00e+00 | 1ARL6IP1 |
150 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
200 |
| HALLMARK_HYPOXIA | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
200 |
| HALLMARK_CHOLESTEROL_HOMEOSTASIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
74 |
| HALLMARK_WNT_BETA_CATENIN_SIGNALING | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
42 |
| HALLMARK_TGF_BETA_SIGNALING | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
54 |
| HALLMARK_IL6_JAK_STAT3_SIGNALING | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
87 |
| HALLMARK_NOTCH_SIGNALING | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
32 |
Dowload full table
KEGG Pathways:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| KEGG_CELL_CYCLE | 9.92e-10 | 31.36 | 12.44 | 1.85e-07 | 1.85e-07 | 8CDK1, CDC20, CCNB1, CCNA2, PLK1, BUB1B, PTTG1, CCNB2 |
125 |
| KEGG_OOCYTE_MEIOSIS | 1.51e-08 | 29.59 | 10.98 | 1.40e-06 | 2.81e-06 | 7CDK1, CDC20, CCNB1, PLK1, AURKA, PTTG1, CCNB2 |
113 |
| KEGG_PROGESTERONE_MEDIATED_OOCYTE_MATURATION | 2.59e-06 | 26.76 | 8.08 | 1.61e-04 | 4.82e-04 | 5CDK1, CCNB1, CCNA2, PLK1, CCNB2 |
85 |
| KEGG_GAP_JUNCTION | 8.44e-05 | 19.48 | 4.98 | 3.93e-03 | 1.57e-02 | 4CDK1, TUBB4B, TUBA1C, TUBA1B |
90 |
| KEGG_PATHOGENIC_ESCHERICHIA_COLI_INFECTION | 4.06e-04 | 23.21 | 4.48 | 1.51e-02 | 7.56e-02 | 3TUBB4B, TUBA1C, TUBA1B |
56 |
| KEGG_P53_SIGNALING_PATHWAY | 7.18e-04 | 18.92 | 3.67 | 2.23e-02 | 1.34e-01 | 3CDK1, CCNB1, CCNB2 |
68 |
| KEGG_UBIQUITIN_MEDIATED_PROTEOLYSIS | 5.04e-03 | 9.32 | 1.83 | 1.34e-01 | 9.38e-01 | 3UBE2C, CDC20, UBE2S |
135 |
| KEGG_COLORECTAL_CANCER | 1.48e-01 | 6.45 | 0.16 | 1.00e+00 | 1.00e+00 | 1BIRC5 |
62 |
| KEGG_ECM_RECEPTOR_INTERACTION | 1.95e-01 | 4.74 | 0.12 | 1.00e+00 | 1.00e+00 | 1HMMR |
84 |
| KEGG_PATHWAYS_IN_CANCER | 5.66e-01 | 1.21 | 0.03 | 1.00e+00 | 1.00e+00 | 1BIRC5 |
325 |
| KEGG_N_GLYCAN_BIOSYNTHESIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
46 |
| KEGG_OTHER_GLYCAN_DEGRADATION | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
16 |
| KEGG_O_GLYCAN_BIOSYNTHESIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
30 |
| KEGG_GLYCOSAMINOGLYCAN_DEGRADATION | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
21 |
| KEGG_GLYCOSAMINOGLYCAN_BIOSYNTHESIS_KERATAN_SULFATE | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
15 |
| KEGG_GLYCEROLIPID_METABOLISM | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
49 |
| KEGG_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI_ANCHOR_BIOSYNTHESIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
25 |
| KEGG_GLYCEROPHOSPHOLIPID_METABOLISM | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
77 |
| KEGG_ETHER_LIPID_METABOLISM | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
33 |
| KEGG_ARACHIDONIC_ACID_METABOLISM | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
58 |
Dowload full table
CHR Positional Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| chr1p34 | 3.30e-02 | 4.52 | 0.90 | 1.00e+00 | 1.00e+00 | 3CDC20, CDCA8, KIF2C |
275 |
| chr15q15 | 5.22e-02 | 5.70 | 0.66 | 1.00e+00 | 1.00e+00 | 2NUSAP1, BUB1B |
143 |
| chr16p12 | 6.63e-02 | 4.96 | 0.58 | 1.00e+00 | 1.00e+00 | 2PLK1, ARL6IP1 |
164 |
| chr10q23 | 8.52e-02 | 4.27 | 0.50 | 1.00e+00 | 1.00e+00 | 2KIF11, KIF20B |
190 |
| chr4q27 | 7.23e-02 | 14.05 | 0.34 | 1.00e+00 | 1.00e+00 | 1CCNA2 |
29 |
| chr5q34 | 9.36e-02 | 10.63 | 0.26 | 1.00e+00 | 1.00e+00 | 1HMMR |
38 |
| chr20q13 | 2.69e-01 | 2.02 | 0.24 | 1.00e+00 | 1.00e+00 | 2UBE2C, AURKA |
400 |
| chr12q13 | 2.76e-01 | 1.98 | 0.23 | 1.00e+00 | 1.00e+00 | 2TUBA1C, TUBA1B |
407 |
| chr4q24 | 1.35e-01 | 7.15 | 0.17 | 1.00e+00 | 1.00e+00 | 1CENPE |
56 |
| chr15q23 | 1.55e-01 | 6.15 | 0.15 | 1.00e+00 | 1.00e+00 | 1KIF23 |
65 |
| chr1q31 | 1.68e-01 | 5.62 | 0.14 | 1.00e+00 | 1.00e+00 | 1ASPM |
71 |
| chr14q22 | 2.08e-01 | 4.42 | 0.11 | 1.00e+00 | 1.00e+00 | 1DLGAP5 |
90 |
| chr14q21 | 2.10e-01 | 4.37 | 0.11 | 1.00e+00 | 1.00e+00 | 1MIS18BP1 |
91 |
| chr1q41 | 2.12e-01 | 4.32 | 0.11 | 1.00e+00 | 1.00e+00 | 1CENPF |
92 |
| chr17q24 | 2.16e-01 | 4.23 | 0.10 | 1.00e+00 | 1.00e+00 | 1KPNA2 |
94 |
| chr5q33 | 2.45e-01 | 3.64 | 0.09 | 1.00e+00 | 1.00e+00 | 1PTTG1 |
109 |
| chr10q21 | 2.59e-01 | 3.42 | 0.08 | 1.00e+00 | 1.00e+00 | 1CDK1 |
116 |
| chr15q22 | 2.74e-01 | 3.20 | 0.08 | 1.00e+00 | 1.00e+00 | 1CCNB2 |
124 |
| chr9q31 | 2.81e-01 | 3.10 | 0.08 | 1.00e+00 | 1.00e+00 | 1SMC2 |
128 |
| chr5q13 | 3.07e-01 | 2.79 | 0.07 | 1.00e+00 | 1.00e+00 | 1CCNB1 |
142 |
Dowload full table
Transcription Factor Targets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| HSD17B8_TARGET_GENES | 7.93e-41 | 72.30 | 38.20 | 8.98e-38 | 8.98e-38 | 35UBE2C, CENPF, CDK1, HMGB2, CDC20, CENPE, BIRC5, TOP2A, PRC1, CENPA, MKI67, CCNA2, KIF23, PLK1, HMMR, CKAP2L, CDCA8, SMC4, TACC3, KPNA2, BUB1B, ASPM, AURKB, KIF20A, MIS18BP1, CDCA3, KIF22, INCENP, PTTG1, DLGAP5, KIF2C, KIF20B, CCNB2, SMC2, SPC25 |
659 |
| BARX2_TARGET_GENES | 8.02e-16 | 12.32 | 6.78 | 4.55e-13 | 9.09e-13 | 26UBE2C, CKS2, CENPF, CDK1, HMGB2, CDC20, TPX2, CCNB1, TOP2A, PRC1, CENPA, KIF23, CDCA8, KPNA2, ASPM, AURKB, MIS18BP1, TUBB4B, UBE2S, CDCA3, ARL6IP1, INCENP, DLGAP5, KIF2C, CCNB2, SPC25 |
1723 |
| CC2D1A_TARGET_GENES | 3.07e-12 | 10.50 | 5.63 | 1.02e-09 | 3.48e-09 | 20CENPF, CDK1, HMGB2, CDC20, CENPE, PRC1, CCNA2, PLK1, AURKA, SMC4, BUB1B, ASPM, MIS18BP1, UBE2S, ARL6IP1, INCENP, PTTG1, TUBA1C, KIF20B, SPC25 |
1245 |
| BARX1_TARGET_GENES | 3.99e-12 | 10.34 | 5.55 | 1.02e-09 | 4.52e-09 | 20UBE2C, CKS2, CENPF, HMGB2, CDC20, CENPE, TPX2, TOP2A, CENPA, PLK1, CDCA8, KPNA2, BUB1B, MIS18BP1, ARL6IP1, INCENP, TUBA1C, DLGAP5, TUBA1B, SPC25 |
1264 |
| E2F5_TARGET_GENES | 4.50e-12 | 10.27 | 5.51 | 1.02e-09 | 5.10e-09 | 20CENPF, HMGB2, TPX2, BIRC5, TOP2A, CENPA, MKI67, CCNA2, PLK1, CDCA8, SMC4, BUB1B, AURKB, MIS18BP1, CDCA3, KIF22, DLGAP5, KIF20B, CCNB2, SPC25 |
1273 |
| ASH1L_TARGET_GENES | 3.91e-11 | 9.02 | 4.84 | 7.39e-09 | 4.43e-08 | 20CKS2, CENPF, CDK1, CENPE, TPX2, CCNB1, TOP2A, CENPA, HMMR, CKAP2L, AURKA, CDCA8, KPNA2, ASPM, TUBB4B, CDCA3, INCENP, DLGAP5, CCNB2, SPC25 |
1446 |
| NFY_C | 4.05e-05 | 10.77 | 3.72 | 4.42e-03 | 4.59e-02 | 6UBE2C, CKS2, KIF23, KIF20A, ARL6IP1, SPC25 |
250 |
| E2F2_TARGET_GENES | 2.90e-08 | 6.79 | 3.54 | 4.69e-06 | 3.28e-05 | 17CENPF, CDK1, HMGB2, CENPE, TPX2, CCNB1, TOP2A, CENPA, CCNA2, KIF11, BUB1B, AURKB, KIF20A, ARL6IP1, INCENP, SMC2, SPC25 |
1481 |
| GATTGGY_NFY_Q6_01 | 5.22e-07 | 6.45 | 3.20 | 7.39e-05 | 5.91e-04 | 14UBE2C, CKS2, CENPF, CDK1, HMGB2, TPX2, TOP2A, KIF23, PLK1, AURKA, TACC3, KIF20A, TUBA1C, SPC25 |
1177 |
| PSMB5_TARGET_GENES | 1.24e-04 | 8.73 | 3.02 | 1.08e-02 | 1.40e-01 | 6CKS2, TUBB4B, UBE2S, ARL6IP1, TUBA1C, TUBA1B |
307 |
| AEBP2_TARGET_GENES | 5.53e-06 | 5.97 | 2.83 | 6.96e-04 | 6.26e-03 | 12UBE2C, CENPF, CDK1, CENPE, TPX2, CCNB1, TOP2A, CENPA, TUBB4B, UBE2S, CDCA3, SPC25 |
1033 |
| RRCCGTTA_UNKNOWN | 1.51e-03 | 14.47 | 2.83 | 8.56e-02 | 1.00e+00 | 3AURKA, UBE2S, TUBA1B |
88 |
| NFY_01 | 5.25e-04 | 8.37 | 2.57 | 3.62e-02 | 5.94e-01 | 5CKS2, CDK1, KIF23, PLK1, TACC3 |
261 |
| NFY_Q6 | 5.43e-04 | 8.30 | 2.55 | 3.62e-02 | 6.15e-01 | 5CKS2, CENPF, CDK1, HMGB2, TOP2A |
263 |
| ALPHACP1_01 | 5.43e-04 | 8.30 | 2.55 | 3.62e-02 | 6.15e-01 | 5UBE2C, CKS2, ASPM, CDCA3, DLGAP5 |
263 |
| PRKDC_TARGET_GENES | 4.68e-05 | 5.57 | 2.48 | 4.42e-03 | 5.30e-02 | 10UBE2C, TOP2A, PRC1, CENPA, TUBB4B, UBE2S, CDCA3, INCENP, DLGAP5, CCNB2 |
875 |
| CEBPZ_TARGET_GENES | 4.32e-05 | 4.50 | 2.19 | 4.42e-03 | 4.90e-02 | 13UBE2C, TOP2A, CENPA, CCNA2, CKAP2L, AURKA, CDCA8, TUBB4B, CDCA3, KIF22, INCENP, DLGAP5, SPC25 |
1520 |
| CHAMP1_TARGET_GENES | 1.96e-03 | 8.22 | 2.13 | 1.01e-01 | 1.00e+00 | 4CENPF, CENPE, CCNA2, MIS18BP1 |
208 |
| TCCCRNNRTGC_UNKNOWN | 2.24e-03 | 7.91 | 2.05 | 1.11e-01 | 1.00e+00 | 4PLK1, KIF11, BUB1B, SPC25 |
216 |
| ZBTB49_TARGET_GENES | 1.29e-02 | 98.09 | 1.96 | 4.29e-01 | 1.00e+00 | 1TUBA1B |
5 |
Dowload full table
GO Biological Processes:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GOBP_MEIOTIC_CHROMOSOME_CONDENSATION | 3.24e-07 | 403.56 | 53.00 | 3.36e-05 | 2.42e-03 | 3SMC4, INCENP, SMC2 |
6 |
| GOBP_CELL_DIVISION | 4.22e-46 | 97.51 | 50.32 | 3.16e-42 | 3.16e-42 | 37UBE2C, CKS2, CENPF, CDK1, CDC20, CENPE, TPX2, CCNB1, BIRC5, TOP2A, NUSAP1, PRC1, CENPA, CCNA2, KIF23, PLK1, AURKA, KIF11, CDCA8, SMC4, TACC3, BUB1B, ASPM, AURKB, KIF20A, MIS18BP1, UBE2S, CDCA3, INCENP, PTTG1, TUBA1C, KIF2C, TUBA1B, KIF20B, CCNB2, SMC2, SPC25 |
600 |
| GOBP_KINETOCHORE_ORGANIZATION | 1.58e-11 | 163.13 | 50.00 | 3.19e-09 | 1.18e-07 | 6CENPF, CENPE, CENPA, SMC4, DLGAP5, SMC2 |
22 |
| GOBP_MITOTIC_NUCLEAR_DIVISION | 1.80e-38 | 91.37 | 49.80 | 6.72e-35 | 1.34e-34 | 28UBE2C, CENPF, CDK1, CDC20, CENPE, TPX2, CCNB1, NUSAP1, PRC1, MKI67, KIF23, PLK1, AURKA, KIF11, CDCA8, SMC4, TACC3, BUB1B, AURKB, UBE2S, KIF22, INCENP, PTTG1, DLGAP5, KIF2C, KIF20B, CCNB2, SMC2 |
296 |
| GOBP_MITOTIC_SISTER_CHROMATID_SEGREGATION | 3.63e-29 | 89.12 | 46.80 | 3.02e-26 | 2.72e-25 | 20UBE2C, CENPF, CDC20, CENPE, CCNB1, NUSAP1, PRC1, KIF23, PLK1, CDCA8, SMC4, TACC3, BUB1B, AURKB, KIF22, INCENP, PTTG1, DLGAP5, KIF2C, SMC2 |
164 |
| GOBP_MITOTIC_NUCLEAR_ENVELOPE_DISASSEMBLY | 1.93e-08 | 207.66 | 44.32 | 2.44e-06 | 1.44e-04 | 4CDK1, CCNB1, PLK1, CCNB2 |
12 |
| GOBP_SPINDLE_MIDZONE_ASSEMBLY | 1.93e-08 | 207.66 | 44.32 | 2.44e-06 | 1.44e-04 | 4PRC1, KIF23, AURKB, INCENP |
12 |
| GOBP_REGULATION_OF_CHROMOSOME_SEGREGATION | 4.88e-20 | 88.89 | 41.69 | 2.03e-17 | 3.65e-16 | 13UBE2C, CENPF, CDC20, CENPE, CCNB1, MKI67, PLK1, TACC3, BUB1B, AURKB, PTTG1, DLGAP5, KIF2C |
89 |
| GOBP_SISTER_CHROMATID_SEGREGATION | 2.63e-29 | 78.35 | 41.57 | 2.46e-26 | 1.97e-25 | 21UBE2C, CENPF, CDC20, CENPE, CCNB1, TOP2A, NUSAP1, PRC1, KIF23, PLK1, CDCA8, SMC4, TACC3, BUB1B, AURKB, KIF22, INCENP, PTTG1, DLGAP5, KIF2C, SMC2 |
199 |
| GOBP_POSITIVE_REGULATION_OF_UBIQUITIN_PROTEIN_LIGASE_ACTIVITY | 2.78e-08 | 184.41 | 40.22 | 3.46e-06 | 2.08e-04 | 4UBE2C, CDC20, PLK1, UBE2S |
13 |
| GOBP_REGULATION_OF_CHROMOSOME_SEPARATION | 2.34e-17 | 90.42 | 39.80 | 8.33e-15 | 1.75e-13 | 11UBE2C, CENPF, CDC20, CENPE, CCNB1, PLK1, TACC3, BUB1B, AURKB, PTTG1, DLGAP5 |
71 |
| GOBP_METAPHASE_ANAPHASE_TRANSITION_OF_CELL_CYCLE | 6.80e-16 | 88.98 | 37.75 | 2.21e-13 | 5.08e-12 | 10UBE2C, CENPF, CDC20, CENPE, CCNB1, PLK1, TACC3, BUB1B, AURKB, DLGAP5 |
64 |
| GOBP_ORGANELLE_FISSION | 6.02e-36 | 63.41 | 34.58 | 9.00e-33 | 4.50e-32 | 30UBE2C, CKS2, CENPF, CDK1, CDC20, CENPE, TPX2, CCNB1, TOP2A, NUSAP1, PRC1, MKI67, KIF23, PLK1, AURKA, KIF11, CDCA8, SMC4, TACC3, BUB1B, AURKB, UBE2S, KIF22, INCENP, PTTG1, DLGAP5, KIF2C, KIF20B, CCNB2, SMC2 |
486 |
| GOBP_MEMBRANE_DISASSEMBLY | 5.28e-08 | 152.08 | 33.97 | 6.37e-06 | 3.95e-04 | 4CDK1, CCNB1, PLK1, CCNB2 |
15 |
| GOBP_CHROMOSOME_SEPARATION | 9.51e-18 | 73.21 | 33.62 | 3.56e-15 | 7.12e-14 | 12UBE2C, CENPF, CDC20, CENPE, CCNB1, TOP2A, PLK1, TACC3, BUB1B, AURKB, PTTG1, DLGAP5 |
95 |
| GOBP_SPINDLE_ELONGATION | 1.35e-06 | 203.48 | 32.05 | 1.26e-04 | 1.01e-02 | 3PRC1, KIF23, AURKB |
9 |
| GOBP_MICROTUBULE_CYTOSKELETON_ORGANIZATION_INVOLVED_IN_MITOSIS | 8.30e-21 | 64.45 | 31.90 | 3.65e-18 | 6.21e-17 | 15CDC20, CENPE, TPX2, CCNB1, NUSAP1, PRC1, CENPA, KIF23, PLK1, AURKA, KIF11, TACC3, AURKB, DLGAP5, SPC25 |
143 |
| GOBP_CHROMOSOME_SEGREGATION | 1.47e-29 | 56.80 | 30.87 | 1.57e-26 | 1.10e-25 | 24UBE2C, CENPF, CDC20, CENPE, CCNB1, BIRC5, TOP2A, NUSAP1, PRC1, MKI67, KIF23, PLK1, CDCA8, SMC4, TACC3, BUB1B, AURKB, KIF22, INCENP, PTTG1, DLGAP5, KIF2C, SMC2, SPC25 |
337 |
| GOBP_MITOTIC_SPINDLE_ORGANIZATION | 2.25e-18 | 64.42 | 30.53 | 8.87e-16 | 1.68e-14 | 13CDC20, CENPE, TPX2, CCNB1, PRC1, KIF23, PLK1, AURKA, KIF11, TACC3, AURKB, DLGAP5, SPC25 |
118 |
| GOBP_NUCLEAR_CHROMOSOME_SEGREGATION | 2.27e-26 | 55.37 | 29.57 | 1.70e-23 | 1.70e-22 | 21UBE2C, CENPF, CDC20, CENPE, CCNB1, TOP2A, NUSAP1, PRC1, KIF23, PLK1, CDCA8, SMC4, TACC3, BUB1B, AURKB, KIF22, INCENP, PTTG1, DLGAP5, KIF2C, SMC2 |
273 |
Dowload full table
Immunological Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GSE15750_DAY6_VS_DAY10_TRAF6KO_EFF_CD8_TCELL_UP | 4.78e-50 | 186.50 | 99.16 | 2.33e-46 | 2.33e-46 | 31UBE2C, CENPF, CDK1, HMGB2, CENPE, TPX2, BIRC5, TOP2A, CENPA, MKI67, CCNA2, KIF23, PLK1, HMMR, CKAP2L, AURKA, TACC3, BUB1B, ASPM, AURKB, KIF20A, MIS18BP1, TUBB4B, CDCA3, KIF22, INCENP, TUBA1B, KIF20B, CCNB2, SMC2, SPC25 |
200 |
| GOLDRATH_EFF_VS_MEMORY_CD8_TCELL_UP | 1.36e-45 | 154.74 | 83.89 | 2.21e-42 | 6.62e-42 | 29CKS2, CDK1, CDC20, BIRC5, TOP2A, NUSAP1, PRC1, CENPA, MKI67, CCNA2, KIF23, AURKA, KIF11, CDCA8, TACC3, KPNA2, ASPM, AURKB, KIF20A, MIS18BP1, CDCA3, KIF22, INCENP, TUBA1C, DLGAP5, KIF2C, TUBA1B, CCNB2, SMC2 |
200 |
| GSE15750_DAY6_VS_DAY10_EFF_CD8_TCELL_UP | 1.36e-45 | 154.74 | 83.89 | 2.21e-42 | 6.62e-42 | 29UBE2C, CKS2, CDK1, CDC20, CENPE, TPX2, BIRC5, TOP2A, NUSAP1, PRC1, CENPA, KIF23, PLK1, HMMR, CKAP2L, AURKA, KIF11, CDCA8, KPNA2, BUB1B, ASPM, AURKB, KIF20A, KIF22, KIF2C, TUBA1B, CCNB2, SMC2, SPC25 |
200 |
| GSE13547_CTRL_VS_ANTI_IGM_STIM_BCELL_12H_UP | 4.02e-42 | 141.07 | 76.05 | 4.90e-39 | 1.96e-38 | 27CENPF, CDK1, HMGB2, CENPE, TPX2, BIRC5, TOP2A, PRC1, MKI67, CCNA2, KIF23, PLK1, HMMR, AURKA, KIF11, CDCA8, TACC3, BUB1B, ASPM, AURKB, MIS18BP1, CDCA3, INCENP, DLGAP5, CCNB2, SMC2, SPC25 |
187 |
| GSE14415_INDUCED_VS_NATURAL_TREG_DN | 2.22e-40 | 134.32 | 72.52 | 2.16e-37 | 1.08e-36 | 26CENPF, CDK1, TPX2, BIRC5, TOP2A, CENPA, MKI67, CCNA2, KIF23, PLK1, HMMR, CKAP2L, KIF11, CDCA8, TACC3, KPNA2, BUB1B, ASPM, MIS18BP1, CDCA3, KIF22, INCENP, DLGAP5, KIF2C, CCNB2, SMC2 |
181 |
| GSE24634_TEFF_VS_TCONV_DAY7_IN_CULTURE_UP | 3.41e-39 | 119.61 | 64.72 | 2.77e-36 | 1.66e-35 | 26UBE2C, CKS2, CENPF, CDK1, HMGB2, CDC20, CENPE, TPX2, CCNB1, NUSAP1, PRC1, CENPA, CCNA2, PLK1, HMMR, AURKA, KIF11, CDCA8, TACC3, ASPM, KIF20A, KIF22, PTTG1, DLGAP5, KIF2C, CCNB2 |
200 |
| GSE13547_2H_VS_12_H_ANTI_IGM_STIM_BCELL_UP | 1.44e-36 | 117.62 | 63.12 | 8.77e-34 | 7.02e-33 | 24CENPF, CDK1, HMGB2, CDC20, CENPE, TPX2, BIRC5, TOP2A, PRC1, MKI67, CCNA2, HMMR, CKAP2L, KIF11, CDCA8, TACC3, BUB1B, ASPM, MIS18BP1, CDCA3, KIF22, INCENP, PTTG1, CCNB2 |
175 |
| GSE14415_NATURAL_TREG_VS_TCONV_DN | 2.94e-36 | 113.78 | 61.12 | 1.59e-33 | 1.43e-32 | 24CENPF, CDK1, HMGB2, CENPE, TPX2, BIRC5, TOP2A, PRC1, CCNA2, KIF23, PLK1, HMMR, CKAP2L, AURKA, KIF11, CDCA8, TACC3, BUB1B, ASPM, MIS18BP1, CDCA3, DLGAP5, CCNB2, SMC2 |
180 |
| GSE30962_PRIMARY_VS_SECONDARY_ACUTE_LCMV_INF_CD8_TCELL_UP | 3.92e-37 | 109.67 | 59.31 | 2.73e-34 | 1.91e-33 | 25CKS2, CENPF, HMGB2, CENPE, BIRC5, TOP2A, NUSAP1, CENPA, CCNA2, KIF23, CKAP2L, KPNA2, BUB1B, ASPM, AURKB, KIF20A, MIS18BP1, TUBB4B, CDCA3, KIF22, DLGAP5, KIF20B, CCNB2, SMC2, SPC25 |
200 |
| GSE13547_CTRL_VS_ANTI_IGM_STIM_BCELL_2H_UP | 4.19e-34 | 102.81 | 55.18 | 1.70e-31 | 2.04e-30 | 23CENPF, CDK1, CENPE, TPX2, TOP2A, PRC1, CENPA, MKI67, CCNA2, PLK1, HMMR, KIF11, CDCA8, KPNA2, ASPM, AURKB, MIS18BP1, CDCA3, KIF22, INCENP, KIF2C, CCNB2, SPC25 |
182 |
| GSE36476_CTRL_VS_TSST_ACT_72H_MEMORY_CD4_TCELL_YOUNG_DN | 4.14e-35 | 101.09 | 54.30 | 1.83e-32 | 2.02e-31 | 24UBE2C, CKS2, CDC20, CENPE, CCNB1, BIRC5, TOP2A, NUSAP1, CENPA, MKI67, HMMR, AURKA, KIF11, CDCA8, BUB1B, ASPM, KIF20A, CDCA3, PTTG1, TUBA1C, DLGAP5, KIF2C, TUBA1B, CCNB2 |
200 |
| GSE36476_CTRL_VS_TSST_ACT_72H_MEMORY_CD4_TCELL_OLD_DN | 4.14e-35 | 101.09 | 54.30 | 1.83e-32 | 2.02e-31 | 24UBE2C, CENPF, CDC20, TPX2, CCNB1, TOP2A, NUSAP1, CENPA, MKI67, CCNA2, KIF23, HMMR, AURKA, KIF11, BUB1B, ASPM, AURKB, KIF20A, CDCA3, TUBA1C, DLGAP5, TUBA1B, CCNB2, SMC2 |
200 |
| GSE45365_HEALTHY_VS_MCMV_INFECTION_CD11B_DC_DN | 2.19e-33 | 95.36 | 51.11 | 8.22e-31 | 1.07e-29 | 23UBE2C, CENPF, CDK1, CDC20, CENPE, TPX2, CCNB1, BIRC5, PRC1, CENPA, MKI67, PLK1, HMMR, AURKA, CDCA8, ASPM, AURKB, KIF20A, UBE2S, CDCA3, PTTG1, DLGAP5, CCNB2 |
195 |
| GOLDRATH_NAIVE_VS_EFF_CD8_TCELL_DN | 4.01e-33 | 92.67 | 49.70 | 1.15e-30 | 1.96e-29 | 23CKS2, CDK1, BIRC5, TOP2A, NUSAP1, PRC1, CENPA, MKI67, CCNA2, AURKA, KIF11, CDCA8, TACC3, ASPM, AURKB, KIF20A, MIS18BP1, CDCA3, KIF22, INCENP, DLGAP5, KIF2C, CCNB2 |
200 |
| GSE24634_TREG_VS_TCONV_POST_DAY7_IL4_CONVERSION_UP | 4.01e-33 | 92.67 | 49.70 | 1.15e-30 | 1.96e-29 | 23UBE2C, CKS2, CENPF, CDC20, CENPE, TPX2, CCNB1, BIRC5, NUSAP1, PRC1, CENPA, CCNA2, AURKA, KIF11, CDCA8, AURKB, TUBB4B, UBE2S, KIF22, PTTG1, DLGAP5, KIF2C, CCNB2 |
200 |
| GSE36476_CTRL_VS_TSST_ACT_40H_MEMORY_CD4_TCELL_YOUNG_DN | 4.01e-33 | 92.67 | 49.70 | 1.15e-30 | 1.96e-29 | 23UBE2C, CKS2, CDC20, CENPE, TPX2, CCNB1, BIRC5, TOP2A, NUSAP1, CENPA, MKI67, KIF23, HMMR, KIF11, CDCA8, ASPM, AURKB, KIF20A, CDCA3, TUBA1C, KIF2C, TUBA1B, CCNB2 |
200 |
| GSE39556_CD8A_DC_VS_NK_CELL_MOUSE_3H_POST_POLYIC_INJ_UP | 4.01e-33 | 92.67 | 49.70 | 1.15e-30 | 1.96e-29 | 23CKS2, CDK1, CDC20, CENPE, TPX2, CCNB1, TOP2A, CENPA, CCNA2, KIF23, HMMR, AURKA, KIF11, SMC4, KPNA2, BUB1B, TUBB4B, UBE2S, KIF22, DLGAP5, KIF2C, KIF20B, CCNB2 |
200 |
| GSE27241_WT_VS_RORGT_KO_TH17_POLARIZED_CD4_TCELL_UP | 3.20e-29 | 89.75 | 47.12 | 8.20e-27 | 1.56e-25 | 20CENPF, CENPE, TPX2, TOP2A, PRC1, MKI67, CCNA2, KIF23, PLK1, CKAP2L, KIF11, CDCA8, TACC3, BUB1B, ASPM, MIS18BP1, UBE2S, KIF22, INCENP, SMC2 |
163 |
| GSE24634_IL4_VS_CTRL_TREATED_NAIVE_CD4_TCELL_DAY7_UP | 2.85e-31 | 85.97 | 45.88 | 7.71e-29 | 1.39e-27 | 22UBE2C, CDK1, HMGB2, CDC20, TPX2, CCNB1, BIRC5, NUSAP1, MKI67, CCNA2, KIF23, HMMR, KIF11, CDCA8, BUB1B, AURKB, KIF20A, TUBB4B, PTTG1, DLGAP5, KIF2C, CCNB2 |
198 |
| GSE2405_HEAT_KILLED_LYSATE_VS_LIVE_A_PHAGOCYTOPHILUM_STIM_NEUTROPHIL_9H_UP | 1.99e-27 | 71.73 | 37.88 | 4.61e-25 | 9.68e-24 | 20CKS2, CENPF, HMGB2, CDC20, CENPE, PRC1, MKI67, HMMR, CKAP2L, AURKA, KIF11, CDCA8, KPNA2, BUB1B, ASPM, KIF20A, KIF2C, TUBA1B, CCNB2, SPC25 |
199 |
Top Ranked Transcription Factors for this Gene Expression Program:
| Gene Symbol | TF Rank | DNA Binding Domain | Motif Status | IUPAC PWM | GTEx | DepMap | Decartes |
|---|---|---|---|---|---|---|---|
| HMGB2 | 5 | No | Unlikely to be sequence specific TF | Low specificity DNA-binding protein | No motif | None | In the alignment, it clusters with other HMG proteins that have not yielded motifs. Evidence exists showing HMGB½ are non-sequence-specific binding proteins (PMID: 11497996). HMGB2 is well-known to facilitate DNA-bending and interact with sequence-specific TFs and transcriptional co-factors. |
| TOP2A | 12 | No | Unlikely to be sequence specific TF | Low specificity DNA-binding protein | No motif | None | DNA topoisomerase. Binds DNA in the crystal structure (PDB: 4FM9). |
| CENPA | 15 | Yes | Likely to be sequence specific TF | Low specificity DNA-binding protein | No motif | None | Histone like protein. Binds DNA in the crystal structure (PDB: 3AN2). |
| MIS18BP1 | 33 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Contains 1 SANT domain, and a SANTA domain |
| PTTG1 | 41 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | GO annotation has no evidence for DNA-binding activity (PMID:9811450), only transcriptional activation in reporter experiments |
| KIF15 | 60 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Has a bZIP like fragment that lacks the basic region required for DNA binding and a STE-domain that is classified as a potential DBD in CIS-BP. It is a kinesin operating in the microtubule system (PMID: 24419385) |
| NDC80 | 63 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Based on the alignments, the protein is only weakly related to the other bZIP proteins and lacks the basic region required for DNA binding |
| NUCKS1 | 77 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Contains a GRP core, which is similar to an AT-hook. But, the amino acids flanking the core are quite different from an AT-hook. Could non-specifically bind to DNA, but lacks compelling evidence. |
| HMGB1 | 80 | No | Unlikely to be sequence specific TF | Low specificity DNA-binding protein | No motif | None | In the alignment, it clusters with other HMG proteins that have not yielded motifs. Evidence exists showing HMGB½ are non-sequence-specific binding proteins (PMID: 11497996). |
| HIST1H1B | 81 | No | Unlikely to be sequence specific TF | Low specificity DNA-binding protein | No motif | None | Linker histone. To my knowledge there has been no demonstration of sequence specificity of linker histones, so I think this one should be called unlikely. |
| RAD21 | 103 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Motifs are suspicious; protein is a component of the cohesin complex and there is no evidence for direct binding to DNA. |
| MXD3 | 107 | Yes | Inferred motif | Obligate heteromer | In vivo/Misc source | None | All three MXD proteins have very similar sequences and should behave accordingly, making heterodimers with at least MAX (PMID:8521822). |
| HIST1H1E | 113 | No | Unlikely to be sequence specific TF | Low specificity DNA-binding protein | No motif | None | Binds DNA as heteromultimeric complex |
| DEPDC1B | 127 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Protein does not have known DBDs and has been shown to function as an inhibitor of a RhoA-based signaling complex (PMID: 25458010) |
| FOXM1 | 134 | Yes | Known motif | Monomer or homomultimer | 100 perc ID - in vitro | None | None |
| H1FX | 145 | No | Unlikely to be sequence specific TF | Low specificity DNA-binding protein | No motif | None | Histone component |
| H1F0 | 146 | No | Unlikely to be sequence specific TF | Low specificity DNA-binding protein | No motif | None | Histone. Likely a false-positive Pfam match based on the similarity of Forkhead domains to H1 domains |
| HIST1H1A | 154 | No | Unlikely to be sequence specific TF | Low specificity DNA-binding protein | No motif | None | Histone component |
| TERF1 | 161 | Yes | Likely to be sequence specific TF | Monomer or homomultimer | No motif | None | Contains 1 Myb domain, and has structural evidence of DNA-binding (PDB: 1IV6). Subunit of Shelterin, a complex that specifically binds telomeres (PMID: 16166375). |
| EME1 | 171 | No | Unlikely to be sequence specific TF | Low specificity DNA-binding protein | No motif | None | Binds DNA in the crystal structure (PDB: 4P0P) in a heterodimeric complex with MUS81. |
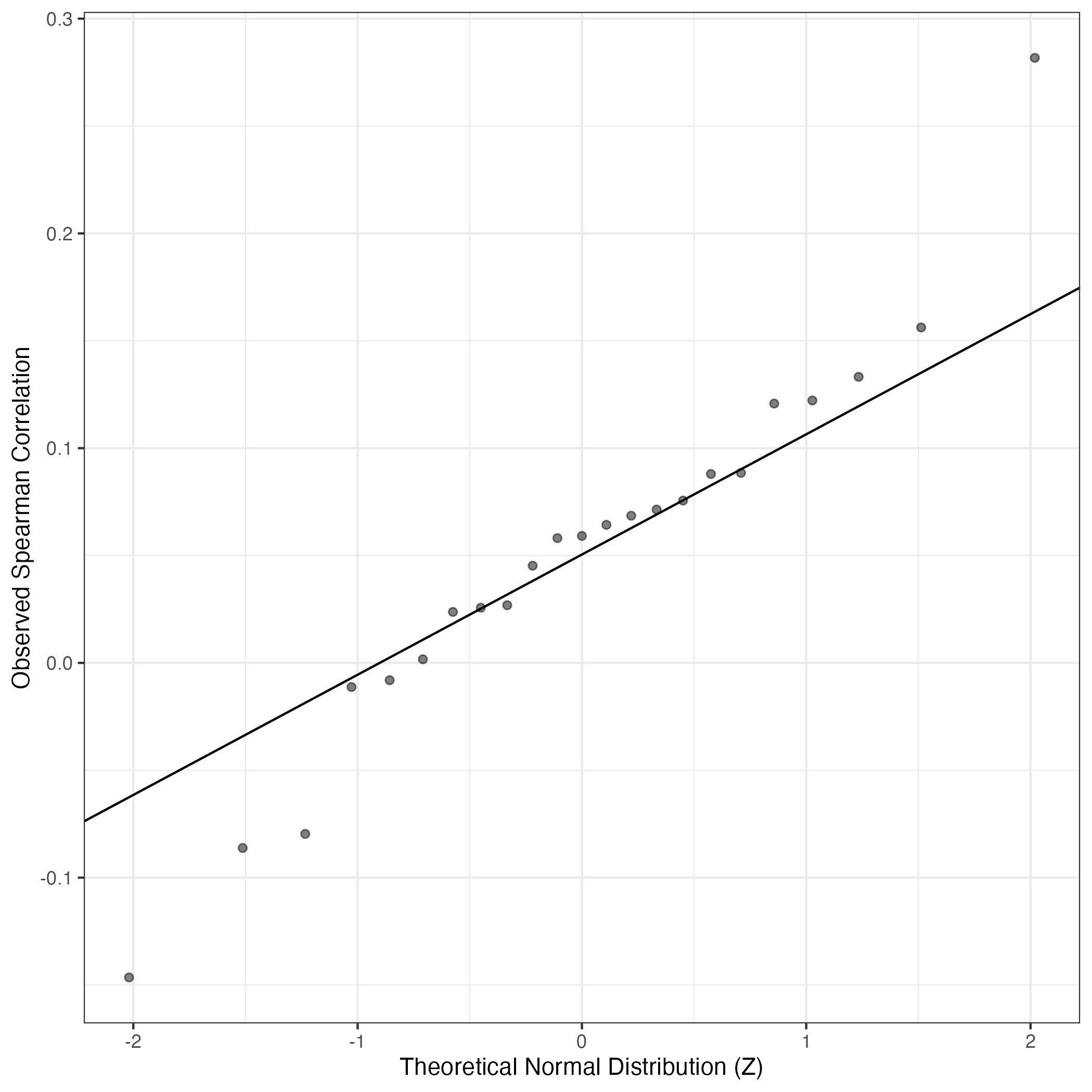
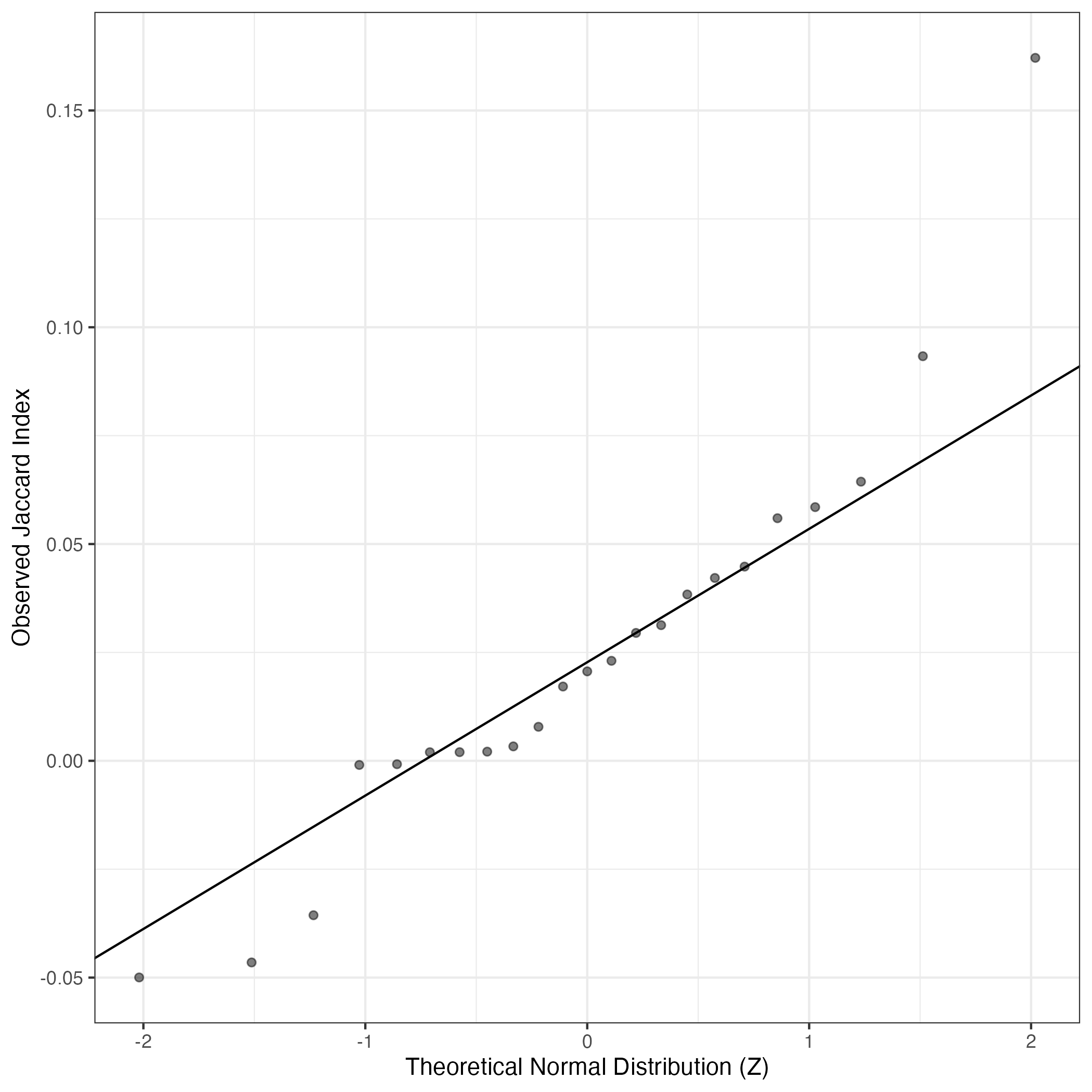
QQ Plot showing correlations with other GEPs in this dataset, calculated by Spearman correlation:
Interactive QQ-plot of gene loadings:
A similar QQ-plot as above, but only for instances where the H value is e.g. > 25, i.e. we are confident that the expression program is active above noise. Agreemenet between these binary vectors is tested using the Jaccard Index, with the P-values calculated by an exact test:
Interactive QQ-plot:
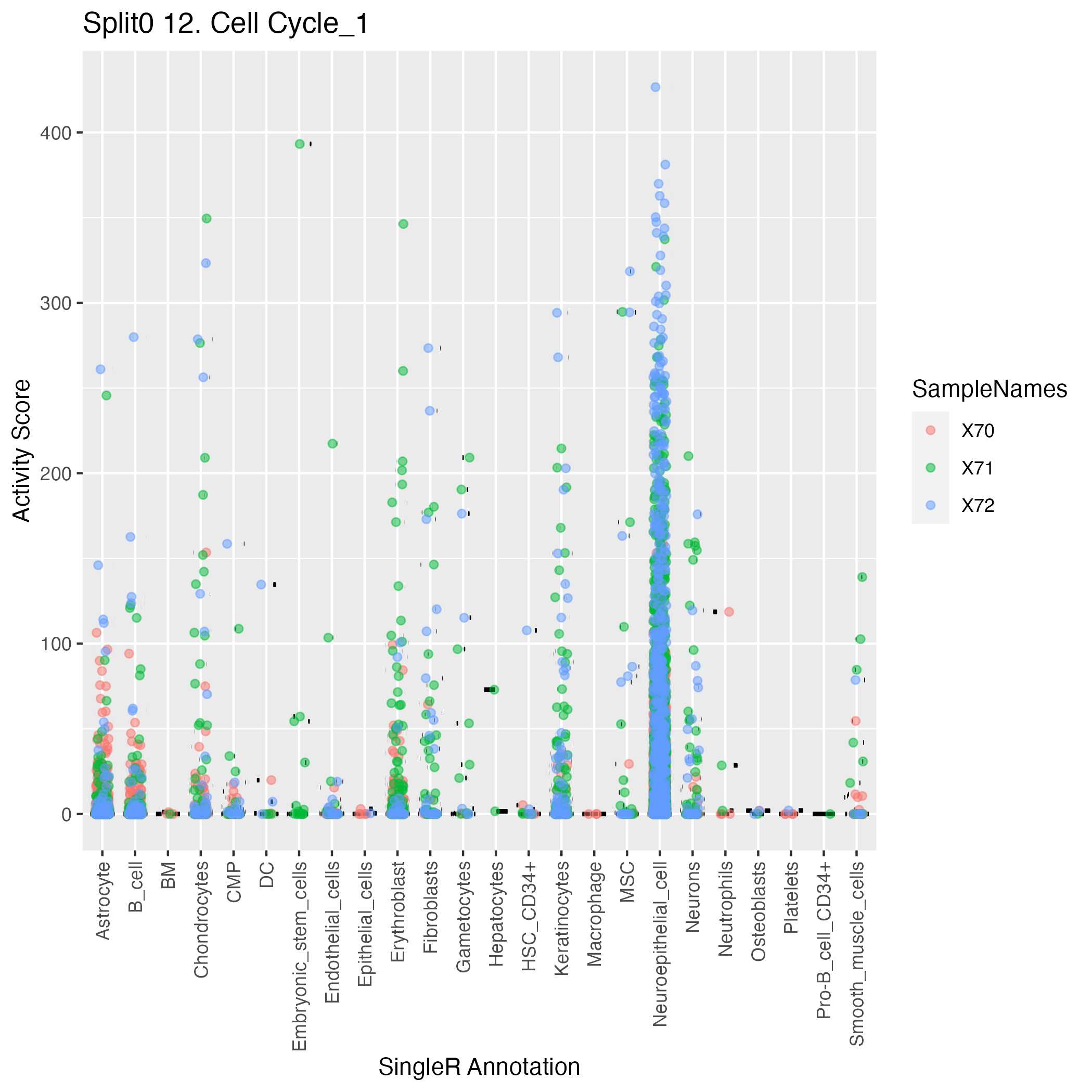
Singler cell type annotations for the top 50 cells on this program.
| Cell ID | Singler label | Singler Delta | Activity Score | Top Singler Raw Scores |
|---|---|---|---|---|
| X72_GGTAATCGTATCGTTG | Neuroepithelial_cell:ESC-derived | 0.37 | 426.56 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.76, Neurons:ES_cell-derived_neural_precursor: 0.76, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.74, MSC: 0.74, Embryonic_stem_cells: 0.72, Keratinocytes: 0.68, Fibroblasts:breast: 0.68, Keratinocytes:IL26: 0.68, iPS_cells:adipose_stem_cells: 0.68, Chondrocytes:MSC-derived: 0.66 |
| X71_TCCCAGTCATTGGCAT | iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff | 0.42 | 393.21 | Raw ScoresiPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.81, Embryonic_stem_cells: 0.78, Neuroepithelial_cell:ESC-derived: 0.75, iPS_cells:fibroblast-derived:Direct_del._reprog: 0.73, Gametocytes:oocyte: 0.71, iPS_cells:fibroblast-derived:Retroviral_transf: 0.71, Astrocyte:Embryonic_stem_cell-derived: 0.69, Keratinocytes:IL26: 0.69, MSC: 0.68, Keratinocytes:KGF: 0.67 |
| X72_AGCGTATTCCAAGCAT | Neuroepithelial_cell:ESC-derived | 0.37 | 381.13 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.76, Neurons:ES_cell-derived_neural_precursor: 0.76, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.74, MSC: 0.74, Embryonic_stem_cells: 0.72, Keratinocytes: 0.68, Fibroblasts:breast: 0.68, Keratinocytes:IL26: 0.68, iPS_cells:adipose_stem_cells: 0.68, Chondrocytes:MSC-derived: 0.66 |
| X72_AAGCATCGTCTAATCG | Keratinocytes:KGF | 0.48 | 369.86 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.42, Keratinocytes:KGF: 0.42, Neurons:ES_cell-derived_neural_precursor: 0.42, Neurons:adrenal_medulla_cell_line: 0.42, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.42, iPS_cells:adipose_stem_cells: 0.42, Chondrocytes:MSC-derived: 0.42, iPS_cells:fibroblast-derived:Direct_del._reprog: 0.41, Erythroblast: 0.4, Gametocytes:oocyte: 0.4 |
| X72_GTCTTTAGTGGCACTC | Neuroepithelial_cell:ESC-derived | 0.32 | 362.78 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.71, Neurons:ES_cell-derived_neural_precursor: 0.68, MSC: 0.67, Keratinocytes:IL26: 0.65, Keratinocytes: 0.65, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.64, Embryonic_stem_cells: 0.64, Chondrocytes:MSC-derived: 0.63, Keratinocytes:IL20: 0.63, Keratinocytes:IFNg: 0.62 |
| X72_AGCCAGCTCATTGCCC | Neuroepithelial_cell:ESC-derived | 0.35 | 358.43 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.58, Neurons:ES_cell-derived_neural_precursor: 0.58, Erythroblast: 0.57, Gametocytes:oocyte: 0.57, MSC: 0.57, Chondrocytes:MSC-derived: 0.54, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.54, Keratinocytes:KGF: 0.5, Neurons:adrenal_medulla_cell_line: 0.5, iPS_cells:adipose_stem_cells: 0.5 |
| X72_CGCCAGAGTGATGGCA | Neurons:ES_cell-derived_neural_precursor | 0.38 | 350.26 | Raw ScoresNeurons:ES_cell-derived_neural_precursor: 0.75, Neuroepithelial_cell:ESC-derived: 0.75, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.75, Embryonic_stem_cells: 0.73, MSC: 0.72, Fibroblasts:breast: 0.7, Keratinocytes: 0.7, Keratinocytes:IL26: 0.7, iPS_cells:adipose_stem_cells: 0.69, Astrocyte:Embryonic_stem_cell-derived: 0.68 |
| X71_TCTCTGGGTCCAATCA | Chondrocytes:MSC-derived | 0.33 | 349.48 | Raw ScoresChondrocytes:MSC-derived: 0.63, iPS_cells:adipose_stem_cells: 0.61, Smooth_muscle_cells:bronchial: 0.58, Smooth_muscle_cells:bronchial:vit_D: 0.58, Fibroblasts:breast: 0.57, MSC: 0.56, Osteoblasts: 0.54, Neurons:ES_cell-derived_neural_precursor: 0.53, Osteoblasts:BMP2: 0.52, Erythroblast: 0.51 |
| X72_CGAAGGACATTGTACG | Neuroepithelial_cell:ESC-derived | 0.37 | 347.39 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.76, Neurons:ES_cell-derived_neural_precursor: 0.76, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.74, MSC: 0.74, Embryonic_stem_cells: 0.72, Keratinocytes: 0.68, Fibroblasts:breast: 0.68, Keratinocytes:IL26: 0.68, iPS_cells:adipose_stem_cells: 0.68, Chondrocytes:MSC-derived: 0.66 |
| X71_ATTTCACCAGAACCGA | Erythroblast | 0.23 | 346.29 | Raw ScoresErythroblast: 0.67, Neuroepithelial_cell:ESC-derived: 0.66, Smooth_muscle_cells:bronchial: 0.65, Neurons:ES_cell-derived_neural_precursor: 0.64, Gametocytes:oocyte: 0.63, MSC: 0.63, Smooth_muscle_cells:bronchial:vit_D: 0.63, Chondrocytes:MSC-derived: 0.62, DC:monocyte-derived:immature: 0.61, Neutrophil:LPS: 0.6 |
| X72_TTCACGCGTCCGAAAG | Neuroepithelial_cell:ESC-derived | 0.33 | 343.72 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.68, Neurons:ES_cell-derived_neural_precursor: 0.68, MSC: 0.65, Chondrocytes:MSC-derived: 0.64, Gametocytes:oocyte: 0.63, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.6, iPS_cells:PDB_2lox-17: 0.59, Erythroblast: 0.59, Embryonic_stem_cells: 0.58, Neurons:adrenal_medulla_cell_line: 0.55 |
| X72_CATCCACTCACCGCTT | Keratinocytes:KGF | 0.48 | 341.08 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.42, Keratinocytes:KGF: 0.42, Neurons:ES_cell-derived_neural_precursor: 0.42, Neurons:adrenal_medulla_cell_line: 0.42, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.42, iPS_cells:adipose_stem_cells: 0.42, Chondrocytes:MSC-derived: 0.42, iPS_cells:fibroblast-derived:Direct_del._reprog: 0.41, Erythroblast: 0.4, Gametocytes:oocyte: 0.4 |
| X72_GGTAACTTCTGCTTAT | iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff | 0.38 | 338.97 | Raw ScoresiPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.75, Neuroepithelial_cell:ESC-derived: 0.75, Neurons:ES_cell-derived_neural_precursor: 0.75, Embryonic_stem_cells: 0.73, MSC: 0.72, Fibroblasts:breast: 0.7, Keratinocytes: 0.7, Keratinocytes:IL26: 0.7, iPS_cells:adipose_stem_cells: 0.7, Astrocyte:Embryonic_stem_cell-derived: 0.68 |
| X71_TAGATCGCATCCTAAG | Neurons:ES_cell-derived_neural_precursor | 0.40 | 337.24 | Raw ScoresNeurons:ES_cell-derived_neural_precursor: 0.68, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.68, Neuroepithelial_cell:ESC-derived: 0.65, Embryonic_stem_cells: 0.65, Keratinocytes:KGF: 0.64, iPS_cells:adipose_stem_cells: 0.64, iPS_cells:fibroblast-derived:Direct_del._reprog: 0.64, Keratinocytes:IL26: 0.63, Fibroblasts:breast: 0.63, Keratinocytes: 0.63 |
| X72_GGGTTTACAGCTAACT | Neuroepithelial_cell:ESC-derived | 0.40 | 327.77 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.54, Neurons:ES_cell-derived_neural_precursor: 0.54, Chondrocytes:MSC-derived: 0.5, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.48, MSC: 0.47, Neurons:adrenal_medulla_cell_line: 0.46, Embryonic_stem_cells: 0.45, iPS_cells:adipose_stem_cells: 0.44, Gametocytes:oocyte: 0.44, Astrocyte:Embryonic_stem_cell-derived: 0.43 |
| X72_AAGACAAGTCAGGTGA | Chondrocytes:MSC-derived | 0.32 | 323.28 | Raw ScoresChondrocytes:MSC-derived: 0.72, MSC: 0.66, iPS_cells:adipose_stem_cells: 0.65, Osteoblasts: 0.63, Smooth_muscle_cells:bronchial: 0.62, Smooth_muscle_cells:bronchial:vit_D: 0.62, Fibroblasts:breast: 0.62, Neuroepithelial_cell:ESC-derived: 0.62, Neurons:ES_cell-derived_neural_precursor: 0.61, Osteoblasts:BMP2: 0.59 |
| X71_TCTATCAAGCACTCGC | Neuroepithelial_cell:ESC-derived | 0.33 | 321.14 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.68, Neurons:ES_cell-derived_neural_precursor: 0.68, MSC: 0.65, Chondrocytes:MSC-derived: 0.64, Gametocytes:oocyte: 0.63, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.6, iPS_cells:PDB_2lox-17: 0.59, Erythroblast: 0.59, Embryonic_stem_cells: 0.58, Neurons:adrenal_medulla_cell_line: 0.55 |
| X72_ATCGTCCCAAGCAGGT | Neuroepithelial_cell:ESC-derived | 0.33 | 319.11 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.69, Neurons:ES_cell-derived_neural_precursor: 0.66, MSC: 0.65, Keratinocytes:IL26: 0.64, Keratinocytes: 0.64, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.62, Astrocyte:Embryonic_stem_cell-derived: 0.62, Embryonic_stem_cells: 0.61, Keratinocytes:IFNg: 0.61, Keratinocytes:IL20: 0.61 |
| X72_AGCGCCAAGCGCCTAC | iPS_cells:CRL2097_foreskin | 0.20 | 318.45 | Raw ScoresiPS_cells:CRL2097_foreskin: 0.57, DC:monocyte-derived:Poly(IC): 0.54, Endothelial_cells:HUVEC:Serum_Amyloid_A: 0.53, Fibroblasts:foreskin: 0.53, iPS_cells:foreskin_fibrobasts: 0.53, Endothelial_cells:HUVEC:IL-1b: 0.52, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.52, iPS_cells:skin_fibroblast: 0.52, MSC: 0.51, Endothelial_cells:lymphatic:KSHV: 0.49 |
| X72_ACTGTGAAGTGCTCGC | Neurons:ES_cell-derived_neural_precursor | 0.29 | 310.14 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.68, Neurons:ES_cell-derived_neural_precursor: 0.68, MSC: 0.66, Gametocytes:oocyte: 0.64, Chondrocytes:MSC-derived: 0.63, iPS_cells:PDB_2lox-17: 0.61, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.6, Erythroblast: 0.59, Embryonic_stem_cells: 0.57, iPS_cells:PDB_1lox-17Puro-10: 0.57 |
| X72_CCACACTTCTGACCCT | Neurons:ES_cell-derived_neural_precursor | 0.27 | 304.45 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.67, Neurons:ES_cell-derived_neural_precursor: 0.67, MSC: 0.65, Chondrocytes:MSC-derived: 0.62, Gametocytes:oocyte: 0.62, iPS_cells:PDB_2lox-17: 0.61, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.58, iPS_cells:PDB_1lox-17Puro-10: 0.58, Embryonic_stem_cells: 0.56, Erythroblast: 0.55 |
| X72_ATCGGCGGTACTCGAT | Neuroepithelial_cell:ESC-derived | 0.35 | 303.76 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.58, Neurons:ES_cell-derived_neural_precursor: 0.58, Erythroblast: 0.57, Gametocytes:oocyte: 0.57, MSC: 0.57, Chondrocytes:MSC-derived: 0.54, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.54, Keratinocytes:KGF: 0.5, Neurons:adrenal_medulla_cell_line: 0.5, iPS_cells:adipose_stem_cells: 0.5 |
| X71_ATTTCTGTCAGCCTCT | Keratinocytes:KGF | 0.48 | 301.62 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.42, Keratinocytes:KGF: 0.42, Neurons:ES_cell-derived_neural_precursor: 0.42, Neurons:adrenal_medulla_cell_line: 0.42, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.42, iPS_cells:adipose_stem_cells: 0.42, Chondrocytes:MSC-derived: 0.42, iPS_cells:fibroblast-derived:Direct_del._reprog: 0.41, Erythroblast: 0.4, Gametocytes:oocyte: 0.4 |
| X72_TCTCACGTCTTTCCGG | Neuroepithelial_cell:ESC-derived | 0.38 | 300.86 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.76, Neurons:ES_cell-derived_neural_precursor: 0.76, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.75, MSC: 0.73, Embryonic_stem_cells: 0.73, Fibroblasts:breast: 0.69, Keratinocytes: 0.69, Keratinocytes:IL26: 0.69, iPS_cells:adipose_stem_cells: 0.69, Astrocyte:Embryonic_stem_cell-derived: 0.67 |
| X72_TCAATTCGTCCAGAAG | Neuroepithelial_cell:ESC-derived | 0.38 | 299.66 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.76, Neurons:ES_cell-derived_neural_precursor: 0.76, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.74, MSC: 0.73, Embryonic_stem_cells: 0.73, Fibroblasts:breast: 0.68, Keratinocytes: 0.68, Keratinocytes:IL26: 0.68, iPS_cells:adipose_stem_cells: 0.68, Keratinocytes:KGF: 0.67 |
| X71_ATCCCTGTCCATTCAT | iPS_cells:adipose_stem_cells | 0.35 | 294.66 | Raw ScoresiPS_cells:adipose_stem_cells: 0.79, Fibroblasts:breast: 0.79, MSC: 0.74, Neurons:ES_cell-derived_neural_precursor: 0.74, Chondrocytes:MSC-derived: 0.73, Neuroepithelial_cell:ESC-derived: 0.72, Tissue_stem_cells:BM_MSC:TGFb3: 0.69, Smooth_muscle_cells:vascular: 0.66, Smooth_muscle_cells:vascular:IL-17: 0.66, Embryonic_stem_cells: 0.66 |
| X72_GGGCGTTCACGCCACA | iPS_cells:adipose_stem_cells | 0.35 | 294.40 | Raw ScoresiPS_cells:adipose_stem_cells: 0.79, Fibroblasts:breast: 0.79, MSC: 0.74, Neurons:ES_cell-derived_neural_precursor: 0.74, Chondrocytes:MSC-derived: 0.73, Neuroepithelial_cell:ESC-derived: 0.72, Tissue_stem_cells:BM_MSC:TGFb3: 0.69, Smooth_muscle_cells:vascular: 0.66, Smooth_muscle_cells:vascular:IL-17: 0.66, Embryonic_stem_cells: 0.66 |
| X72_CACTGGGAGATTGACA | iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff | 0.40 | 294.14 | Raw ScoresiPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.68, Neurons:ES_cell-derived_neural_precursor: 0.68, Keratinocytes:KGF: 0.65, iPS_cells:adipose_stem_cells: 0.65, iPS_cells:fibroblast-derived:Direct_del._reprog: 0.65, Fibroblasts:breast: 0.65, Keratinocytes: 0.65, Keratinocytes:IL19: 0.65, Keratinocytes:IL20: 0.65, Keratinocytes:IL22: 0.65 |
| X72_AACGTCATCCAACTGA | Neuroepithelial_cell:ESC-derived | 0.40 | 293.05 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.54, Neurons:ES_cell-derived_neural_precursor: 0.54, Chondrocytes:MSC-derived: 0.5, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.48, MSC: 0.47, Neurons:adrenal_medulla_cell_line: 0.46, Embryonic_stem_cells: 0.45, iPS_cells:adipose_stem_cells: 0.44, Gametocytes:oocyte: 0.44, Astrocyte:Embryonic_stem_cell-derived: 0.43 |
| X72_GTCATCCTCGCCAACG | iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff | 0.38 | 290.58 | Raw ScoresiPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.75, Neuroepithelial_cell:ESC-derived: 0.75, Neurons:ES_cell-derived_neural_precursor: 0.75, Embryonic_stem_cells: 0.74, MSC: 0.72, Keratinocytes: 0.7, Keratinocytes:IL26: 0.7, Fibroblasts:breast: 0.7, iPS_cells:adipose_stem_cells: 0.7, Astrocyte:Embryonic_stem_cell-derived: 0.68 |
| X72_AAGTCGTCATCGGCCA | Neurons:ES_cell-derived_neural_precursor | 0.33 | 286.13 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.68, Neurons:ES_cell-derived_neural_precursor: 0.68, MSC: 0.64, Chondrocytes:MSC-derived: 0.63, Gametocytes:oocyte: 0.62, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.6, iPS_cells:PDB_2lox-17: 0.59, Erythroblast: 0.57, Embryonic_stem_cells: 0.57, iPS_cells:PDB_1lox-17Puro-10: 0.55 |
| X72_GGTCTGGTCCCAAGTA | Neurons:ES_cell-derived_neural_precursor | 0.25 | 284.42 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.67, Neurons:ES_cell-derived_neural_precursor: 0.67, MSC: 0.66, Gametocytes:oocyte: 0.63, iPS_cells:PDB_2lox-17: 0.62, Chondrocytes:MSC-derived: 0.62, iPS_cells:PDB_1lox-17Puro-10: 0.59, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.58, Erythroblast: 0.56, iPS_cells:PDB_2lox-21: 0.56 |
| X72_GACTCAAGTTTAGAGA | Monocyte | 0.00 | 279.85 | Raw ScoresMonocyte: 0.42, T_cell:CD4+: 0.42, B_cell: 0.42, B_cell:CXCR4+_centroblast: 0.42, B_cell:CXCR4-_centrocyte: 0.42, B_cell:Germinal_center: 0.42, B_cell:Memory: 0.42, B_cell:Naive: 0.42, B_cell:Plasma_cell: 0.42, B_cell:immature: 0.42 |
| X72_TGGATCAAGGATCATA | Keratinocytes:KGF | 0.48 | 279.65 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.42, Keratinocytes:KGF: 0.42, Neurons:ES_cell-derived_neural_precursor: 0.42, Neurons:adrenal_medulla_cell_line: 0.42, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.42, iPS_cells:adipose_stem_cells: 0.42, Chondrocytes:MSC-derived: 0.42, iPS_cells:fibroblast-derived:Direct_del._reprog: 0.41, Erythroblast: 0.4, Gametocytes:oocyte: 0.4 |
| X72_TTCGGTCGTGTACATC | Chondrocytes:MSC-derived | 0.34 | 278.59 | Raw ScoresChondrocytes:MSC-derived: 0.64, iPS_cells:adipose_stem_cells: 0.62, Smooth_muscle_cells:bronchial: 0.58, Fibroblasts:breast: 0.58, Smooth_muscle_cells:bronchial:vit_D: 0.58, MSC: 0.55, Osteoblasts: 0.54, Osteoblasts:BMP2: 0.52, Neurons:ES_cell-derived_neural_precursor: 0.52, Erythroblast: 0.5 |
| X71_TAACTTCAGCATCCTA | Neuroepithelial_cell:ESC-derived | 0.28 | 278.48 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.59, Neurons:ES_cell-derived_neural_precursor: 0.56, DC:monocyte-derived:immature: 0.54, Chondrocytes:MSC-derived: 0.54, Gametocytes:oocyte: 0.53, Erythroblast: 0.53, MSC: 0.52, iPS_cells:PDB_2lox-17: 0.48, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.47, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.47 |
| X72_TCTGTCGTCAACCTTT | Neuroepithelial_cell:ESC-derived | 0.40 | 277.93 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.54, Neurons:ES_cell-derived_neural_precursor: 0.54, Chondrocytes:MSC-derived: 0.5, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.48, MSC: 0.47, Neurons:adrenal_medulla_cell_line: 0.46, Embryonic_stem_cells: 0.45, iPS_cells:adipose_stem_cells: 0.44, Gametocytes:oocyte: 0.44, Astrocyte:Embryonic_stem_cell-derived: 0.43 |
| X72_CATCGGGGTCCGACGT | Neurons:ES_cell-derived_neural_precursor | 0.33 | 276.46 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.68, Neurons:ES_cell-derived_neural_precursor: 0.68, MSC: 0.64, Chondrocytes:MSC-derived: 0.63, Gametocytes:oocyte: 0.62, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.6, iPS_cells:PDB_2lox-17: 0.59, Erythroblast: 0.57, Embryonic_stem_cells: 0.57, iPS_cells:PDB_1lox-17Puro-10: 0.55 |
| X71_GCAACCGAGGGAGGTG | Chondrocytes:MSC-derived | 0.31 | 276.39 | Raw ScoresChondrocytes:MSC-derived: 0.72, MSC: 0.66, iPS_cells:adipose_stem_cells: 0.66, Osteoblasts: 0.65, Smooth_muscle_cells:bronchial: 0.64, Smooth_muscle_cells:bronchial:vit_D: 0.64, Fibroblasts:breast: 0.63, Neuroepithelial_cell:ESC-derived: 0.62, Neurons:ES_cell-derived_neural_precursor: 0.62, Osteoblasts:BMP2: 0.61 |
| X71_TGTAACGGTTAACCTG | Neuroepithelial_cell:ESC-derived | 0.35 | 274.84 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.58, Neurons:ES_cell-derived_neural_precursor: 0.58, Erythroblast: 0.57, Gametocytes:oocyte: 0.57, MSC: 0.57, Chondrocytes:MSC-derived: 0.54, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.54, Keratinocytes:KGF: 0.5, Neurons:adrenal_medulla_cell_line: 0.5, iPS_cells:adipose_stem_cells: 0.5 |
| X72_TATTGGGTCGTCACCT | Fibroblasts:breast | 0.46 | 273.42 | Raw ScoresFibroblasts:breast: 0.59, iPS_cells:adipose_stem_cells: 0.57, Chondrocytes:MSC-derived: 0.5, Tissue_stem_cells:BM_MSC:TGFb3: 0.47, Epithelial_cells:bronchial: 0.44, iPS_cells:fibroblast-derived:Direct_del._reprog: 0.43, DC:monocyte-derived:anti-DC-SIGN_2h: 0.43, Keratinocytes:IL20: 0.43, Keratinocytes:IL26: 0.43, Keratinocytes:IL22: 0.43 |
| X72_CTCCAACAGGTTGCCC | Neuroepithelial_cell:ESC-derived | 0.40 | 268.87 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.54, Neurons:ES_cell-derived_neural_precursor: 0.54, Chondrocytes:MSC-derived: 0.5, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.48, MSC: 0.47, Neurons:adrenal_medulla_cell_line: 0.46, Embryonic_stem_cells: 0.45, iPS_cells:adipose_stem_cells: 0.44, Gametocytes:oocyte: 0.44, Astrocyte:Embryonic_stem_cell-derived: 0.43 |
| X72_CAACAGTCACAATGCT | Neuroepithelial_cell:ESC-derived | 0.35 | 268.30 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.58, Neurons:ES_cell-derived_neural_precursor: 0.58, Erythroblast: 0.57, Gametocytes:oocyte: 0.57, MSC: 0.57, Chondrocytes:MSC-derived: 0.54, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.54, Keratinocytes:KGF: 0.5, Neurons:adrenal_medulla_cell_line: 0.5, iPS_cells:adipose_stem_cells: 0.5 |
| X71_CGAGTGCTCCTCACGT | Neuroepithelial_cell:ESC-derived | 0.38 | 268.19 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.76, Neurons:ES_cell-derived_neural_precursor: 0.76, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.74, MSC: 0.73, Embryonic_stem_cells: 0.73, Fibroblasts:breast: 0.68, Keratinocytes: 0.68, Keratinocytes:IL26: 0.68, iPS_cells:adipose_stem_cells: 0.68, Keratinocytes:KGF: 0.67 |
| X72_CTGAGCGCAGACCCGT | iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff | 0.40 | 268.02 | Raw ScoresiPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.68, Neurons:ES_cell-derived_neural_precursor: 0.68, Keratinocytes:KGF: 0.65, iPS_cells:adipose_stem_cells: 0.65, iPS_cells:fibroblast-derived:Direct_del._reprog: 0.65, Fibroblasts:breast: 0.65, Keratinocytes: 0.65, Keratinocytes:IL19: 0.65, Keratinocytes:IL20: 0.65, Keratinocytes:IL22: 0.65 |
| X71_ATCATTCCACACAGAG | Neuroepithelial_cell:ESC-derived | 0.33 | 267.92 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.68, Neurons:ES_cell-derived_neural_precursor: 0.68, MSC: 0.65, Chondrocytes:MSC-derived: 0.64, Gametocytes:oocyte: 0.63, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.6, iPS_cells:PDB_2lox-17: 0.59, Erythroblast: 0.59, Embryonic_stem_cells: 0.58, Neurons:adrenal_medulla_cell_line: 0.55 |
| X72_GAGAGGTTCCCTTCCC | Neuroepithelial_cell:ESC-derived | 0.35 | 265.71 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.58, Neurons:ES_cell-derived_neural_precursor: 0.58, Erythroblast: 0.57, Gametocytes:oocyte: 0.57, MSC: 0.57, Chondrocytes:MSC-derived: 0.54, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.54, Keratinocytes:KGF: 0.5, Neurons:adrenal_medulla_cell_line: 0.5, iPS_cells:adipose_stem_cells: 0.5 |
| X72_TTCATGTTCAAATAGG | Keratinocytes:KGF | 0.48 | 264.89 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.42, Keratinocytes:KGF: 0.42, Neurons:ES_cell-derived_neural_precursor: 0.42, Neurons:adrenal_medulla_cell_line: 0.42, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.42, iPS_cells:adipose_stem_cells: 0.42, Chondrocytes:MSC-derived: 0.42, iPS_cells:fibroblast-derived:Direct_del._reprog: 0.41, Erythroblast: 0.4, Gametocytes:oocyte: 0.4 |
| X72_TGTCAGACAAAGGCGT | Neuroepithelial_cell:ESC-derived | 0.33 | 262.89 | Raw ScoresNeuroepithelial_cell:ESC-derived: 0.68, Neurons:ES_cell-derived_neural_precursor: 0.68, MSC: 0.65, Chondrocytes:MSC-derived: 0.64, Gametocytes:oocyte: 0.62, iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.6, iPS_cells:PDB_2lox-17: 0.59, Erythroblast: 0.58, Embryonic_stem_cells: 0.57, Neurons:adrenal_medulla_cell_line: 0.55 |
| X72_AGGGAGTAGATCGGTG | iPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff | 0.41 | 260.98 | Raw ScoresiPS_cells:CRL2097_foreskin-derived:d20_hepatic_diff: 0.64, Astrocyte:Embryonic_stem_cell-derived: 0.62, Fibroblasts:breast: 0.62, Keratinocytes: 0.62, Keratinocytes:IL26: 0.62, Neuroepithelial_cell:ESC-derived: 0.61, Neurons:ES_cell-derived_neural_precursor: 0.61, Embryonic_stem_cells: 0.61, iPS_cells:adipose_stem_cells: 0.61, Keratinocytes:KGF: 0.59 |
Below shows the significant enrichments of this GEP for literature curated gene lists
This data was procured from existing single cell RNA-seq maps of neuroblastoma or related relevant data.
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by the P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Proliferating Cells
These are canonical marker genes of cycling cells, obtained from Fig. 3 of Hsiao et al. (PMID 32312741):
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.69e-04
Mean rank of genes in gene set: 8.25
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| UBE2C | 0.0182446 | 1 | GTEx | DepMap | Descartes | 26.85 | 473.51 |
| CDK1 | 0.0152023 | 4 | GTEx | DepMap | Descartes | 9.49 | 71.41 |
| TOP2A | 0.0138475 | 12 | GTEx | DepMap | Descartes | 25.38 | 128.60 |
| MKI67 | 0.0133305 | 16 | GTEx | DepMap | Descartes | 13.00 | 72.38 |
Adrenergic Fig 1D (Olsen)
Similar to above, but for selected adrenergic marker genes:
Wilcoxon ranksum test P-value for gene set overrepresentation: 7.23e-03
Mean rank of genes in gene set: 5726.5
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| RTN1 | 0.0003576 | 2802 | GTEx | DepMap | Descartes | 3.88 | 2.46 |
| ISL1 | 0.0003072 | 3137 | GTEx | DepMap | Descartes | 1.70 | 22.60 |
| TH | 0.0002305 | 3805 | GTEx | DepMap | Descartes | 0.44 | 1.87 |
| MLLT11 | 0.0001825 | 4352 | GTEx | DepMap | Descartes | 1.61 | 14.27 |
| ELAVL4 | 0.0001778 | 4430 | GTEx | DepMap | Descartes | 1.15 | 0.90 |
| DBH | 0.0001494 | 4822 | GTEx | DepMap | Descartes | 0.94 | 9.71 |
| STMN2 | 0.0001451 | 4891 | GTEx | DepMap | Descartes | 4.88 | 10.33 |
| BEX1 | 0.0001393 | 4980 | GTEx | DepMap | Descartes | 4.42 | 307.28 |
| UCHL1 | 0.0001321 | 5114 | GTEx | DepMap | Descartes | 4.40 | 53.91 |
| MAP1B | 0.0000098 | 8036 | GTEx | DepMap | Descartes | 7.21 | 9.86 |
| NRG1 | -0.0000505 | 10182 | GTEx | DepMap | Descartes | 0.24 | 0.02 |
| RGS5 | -0.0001116 | 12167 | GTEx | DepMap | Descartes | 0.15 | 0.44 |
Adrenergic proliferating cluster (Olsen)
Stated on pages 6 and 7 of the main text of Olsen et al. https://www.biorxiv.org/content/10.1101/2020.05.04.077057v1 - it is argued that upon reanalysis of the adrenergic neuroblastoma cells, there is further heterogeneity, grouped into proliferating, mature and immature cell populations. These genes were found as markers for proliferating adrenergic cells:
Wilcoxon ranksum test P-value for gene set overrepresentation: 7.69e-03
Mean rank of genes in gene set: 104
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| MKI67 | 0.0133305 | 16 | GTEx | DepMap | Descartes | 13.00 | 72.38 |
| EZH2 | 0.0038827 | 192 | GTEx | DepMap | Descartes | 7.15 | 15.18 |
Below shows ranks on this GEP for literature curated gene lists for large gene sets
These include those reported as mesenchymal/adrenergic by Van Groningen et al.
High ranks indicate this gene is a driver of this GEP (note these results are not ordered).
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
VanGroningen Adrenergic Genes
Adrenergic marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 8.72e-13
Mean rank of genes in gene set: 7555.64
Median rank of genes in gene set: 6210
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| UBE2C | 0.0182446 | 1 | GTEx | DepMap | Descartes | 26.85 | 473.51 |
| BIRC5 | 0.0138758 | 11 | GTEx | DepMap | Descartes | 13.71 | 338.97 |
| NUSAP1 | 0.0137677 | 13 | GTEx | DepMap | Descartes | 6.87 | 35.45 |
| PRC1 | 0.0137612 | 14 | GTEx | DepMap | Descartes | 7.82 | 57.16 |
| TUBB4B | 0.0111140 | 34 | GTEx | DepMap | Descartes | 23.69 | 1585.68 |
| ARL6IP1 | 0.0110511 | 37 | GTEx | DepMap | Descartes | 24.32 | 376.73 |
| KNSTRN | 0.0091906 | 58 | GTEx | DepMap | Descartes | 3.45 | 13.64 |
| KIF15 | 0.0089686 | 60 | GTEx | DepMap | Descartes | 3.26 | 6.53 |
| PBK | 0.0077547 | 78 | GTEx | DepMap | Descartes | 3.13 | 38.38 |
| NUF2 | 0.0073051 | 88 | GTEx | DepMap | Descartes | 2.02 | 10.08 |
| CDKN3 | 0.0062299 | 114 | GTEx | DepMap | Descartes | 0.87 | 14.02 |
| FOXM1 | 0.0054263 | 134 | GTEx | DepMap | Descartes | 1.12 | 12.47 |
| LSM3 | 0.0051137 | 143 | GTEx | DepMap | Descartes | 7.04 | 133.21 |
| H1FX | 0.0050693 | 145 | GTEx | DepMap | Descartes | 11.28 | 1528.96 |
| HN1 | 0.0047905 | 155 | GTEx | DepMap | Descartes | 20.64 | NA |
| CDCA5 | 0.0043959 | 165 | GTEx | DepMap | Descartes | 1.17 | 25.27 |
| CDKN2C | 0.0039234 | 190 | GTEx | DepMap | Descartes | 0.32 | 7.61 |
| RANBP1 | 0.0037509 | 197 | GTEx | DepMap | Descartes | 21.85 | 290.26 |
| UBE2T | 0.0036587 | 203 | GTEx | DepMap | Descartes | 0.95 | 10.20 |
| ZWILCH | 0.0033578 | 220 | GTEx | DepMap | Descartes | 0.71 | 2.72 |
| RFC4 | 0.0030919 | 244 | GTEx | DepMap | Descartes | 3.17 | 31.81 |
| GMNN | 0.0030779 | 248 | GTEx | DepMap | Descartes | 4.30 | 51.92 |
| RRM2 | 0.0030747 | 249 | GTEx | DepMap | Descartes | 5.15 | 111.29 |
| VRK1 | 0.0030281 | 255 | GTEx | DepMap | Descartes | 1.91 | 3.51 |
| DNAJC9 | 0.0029111 | 266 | GTEx | DepMap | Descartes | 4.91 | 149.86 |
| CSE1L | 0.0027458 | 275 | GTEx | DepMap | Descartes | 2.50 | 7.52 |
| FIGNL1 | 0.0024524 | 309 | GTEx | DepMap | Descartes | 1.33 | 7.36 |
| LSM4 | 0.0024455 | 312 | GTEx | DepMap | Descartes | 10.86 | 238.59 |
| HNRNPA0 | 0.0021327 | 362 | GTEx | DepMap | Descartes | 16.05 | 748.77 |
| CENPU | 0.0021096 | 364 | GTEx | DepMap | Descartes | 0.66 | 2.99 |
VanGroningen Mesenchymal Genes
Mesenchymal marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 13359.41
Median rank of genes in gene set: 15879
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TUBB6 | 0.0024515 | 310 | GTEx | DepMap | Descartes | 1.65 | 22.50 |
| EMP1 | 0.0023445 | 327 | GTEx | DepMap | Descartes | 4.11 | 22.74 |
| HIST1H2BK | 0.0016396 | 503 | GTEx | DepMap | Descartes | 0.21 | 64.48 |
| SEPT10 | 0.0014455 | 599 | GTEx | DepMap | Descartes | 1.33 | 2.13 |
| HSP90B1 | 0.0014237 | 611 | GTEx | DepMap | Descartes | 24.51 | 220.96 |
| PRDX4 | 0.0013236 | 676 | GTEx | DepMap | Descartes | 6.52 | 57.17 |
| CLIC4 | 0.0013110 | 683 | GTEx | DepMap | Descartes | 2.41 | 5.05 |
| PTBP1 | 0.0009186 | 1066 | GTEx | DepMap | Descartes | 7.84 | 93.69 |
| ELAVL1 | 0.0009116 | 1076 | GTEx | DepMap | Descartes | 5.55 | 17.52 |
| CBFB | 0.0008955 | 1111 | GTEx | DepMap | Descartes | 1.78 | 4.73 |
| DUSP6 | 0.0007674 | 1309 | GTEx | DepMap | Descartes | 1.20 | 35.45 |
| ACTN1 | 0.0007455 | 1356 | GTEx | DepMap | Descartes | 2.44 | 3.37 |
| PLSCR1 | 0.0006942 | 1488 | GTEx | DepMap | Descartes | 0.39 | 2.48 |
| RBMS1 | 0.0006554 | 1580 | GTEx | DepMap | Descartes | 5.47 | 3.10 |
| SUCLG2 | 0.0006096 | 1706 | GTEx | DepMap | Descartes | 2.73 | 1.18 |
| MOB1A | 0.0006060 | 1715 | GTEx | DepMap | Descartes | 1.50 | 9.93 |
| HIST1H2AC | 0.0005490 | 1901 | GTEx | DepMap | Descartes | 0.03 | 1.80 |
| MYL12A | 0.0005446 | 1921 | GTEx | DepMap | Descartes | 12.72 | 159.32 |
| RAP1A | 0.0005011 | 2081 | GTEx | DepMap | Descartes | 2.38 | 4.03 |
| SLC38A2 | 0.0004924 | 2127 | GTEx | DepMap | Descartes | 3.43 | 36.80 |
| POLR2L | 0.0004794 | 2176 | GTEx | DepMap | Descartes | 1.84 | 63.77 |
| FAM120A | 0.0004708 | 2207 | GTEx | DepMap | Descartes | 2.23 | 2.90 |
| SPRED1 | 0.0004667 | 2224 | GTEx | DepMap | Descartes | 1.86 | 4.06 |
| ELK4 | 0.0004597 | 2260 | GTEx | DepMap | Descartes | 0.87 | 4.07 |
| ETS1 | 0.0004512 | 2293 | GTEx | DepMap | Descartes | 0.63 | 0.75 |
| MBD2 | 0.0004482 | 2308 | GTEx | DepMap | Descartes | 2.89 | 5.69 |
| SNAI2 | 0.0004325 | 2383 | GTEx | DepMap | Descartes | 1.83 | 64.86 |
| SPRY1 | 0.0004269 | 2413 | GTEx | DepMap | Descartes | 1.85 | 40.44 |
| ANXA1 | 0.0004268 | 2414 | GTEx | DepMap | Descartes | 2.38 | 12.42 |
| IQGAP2 | 0.0004066 | 2535 | GTEx | DepMap | Descartes | 0.68 | 0.43 |
Descartes adrenocortical markers
Top 50 marker genes of adrenocortical cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/adrenocortical/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 7.89e-01
Mean rank of genes in gene set: 10355.12
Median rank of genes in gene set: 11002
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| LDLR | 0.0013689 | 650 | GTEx | DepMap | Descartes | 0.74 | 3.80 |
| HSPD1 | 0.0009582 | 1012 | GTEx | DepMap | Descartes | 20.18 | 222.54 |
| HSPE1 | 0.0007233 | 1409 | GTEx | DepMap | Descartes | 31.97 | 1138.16 |
| HMGCS1 | 0.0007111 | 1436 | GTEx | DepMap | Descartes | 4.58 | 33.95 |
| HMGCR | 0.0006511 | 1595 | GTEx | DepMap | Descartes | 2.27 | 14.89 |
| CYB5B | 0.0005935 | 1752 | GTEx | DepMap | Descartes | 1.63 | 5.07 |
| FDPS | 0.0005828 | 1788 | GTEx | DepMap | Descartes | 3.94 | 56.72 |
| MSMO1 | 0.0005229 | 2010 | GTEx | DepMap | Descartes | 1.95 | 17.66 |
| IGF1R | 0.0004033 | 2551 | GTEx | DepMap | Descartes | 3.47 | 1.49 |
| DHCR24 | 0.0003510 | 2851 | GTEx | DepMap | Descartes | 1.57 | 6.63 |
| MC2R | 0.0002466 | 3654 | GTEx | DepMap | Descartes | 0.02 | 0.08 |
| FDXR | 0.0002188 | 3911 | GTEx | DepMap | Descartes | 0.43 | 6.78 |
| JAKMIP2 | 0.0001693 | 4534 | GTEx | DepMap | Descartes | 0.16 | 0.15 |
| SLC1A2 | 0.0001287 | 5172 | GTEx | DepMap | Descartes | 0.16 | 0.19 |
| FRMD5 | 0.0000684 | 6342 | GTEx | DepMap | Descartes | 0.05 | 0.03 |
| TM7SF2 | 0.0000526 | 6743 | GTEx | DepMap | Descartes | 0.25 | 5.22 |
| STAR | 0.0000445 | 6946 | GTEx | DepMap | Descartes | 0.04 | 0.56 |
| DNER | 0.0000219 | 7628 | GTEx | DepMap | Descartes | 0.28 | 0.13 |
| DHCR7 | 0.0000217 | 7632 | GTEx | DepMap | Descartes | 0.87 | 4.53 |
| SGCZ | -0.0000028 | 8486 | GTEx | DepMap | Descartes | 0.02 | 0.00 |
| CYP11A1 | -0.0000282 | 9393 | GTEx | DepMap | Descartes | 0.03 | 0.15 |
| POR | -0.0001263 | 12611 | GTEx | DepMap | Descartes | 1.17 | 2.45 |
| SH3BP5 | -0.0001393 | 12993 | GTEx | DepMap | Descartes | 0.83 | 1.34 |
| CYP11B1 | -0.0001636 | 13644 | GTEx | DepMap | Descartes | 0.02 | 0.25 |
| ERN1 | -0.0002080 | 14677 | GTEx | DepMap | Descartes | 0.13 | 0.15 |
| SLC16A9 | -0.0002526 | 15445 | GTEx | DepMap | Descartes | 0.03 | 0.11 |
| FDX1 | -0.0002713 | 15737 | GTEx | DepMap | Descartes | 1.82 | 10.54 |
| PAPSS2 | -0.0002991 | 16135 | GTEx | DepMap | Descartes | 0.55 | 1.50 |
| GRAMD1B | -0.0002998 | 16139 | GTEx | DepMap | Descartes | 0.15 | 0.10 |
| SCAP | -0.0003153 | 16352 | GTEx | DepMap | Descartes | 0.77 | 1.76 |
Descartes chromaffin markers
Top 50 marker genes of chromaffin cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/chromaffin/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 4.96e-02
Mean rank of genes in gene set: 8280.02
Median rank of genes in gene set: 7946.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| MAB21L1 | 0.0006945 | 1487 | GTEx | DepMap | Descartes | 0.32 | 22.72 |
| TUBA1A | 0.0005872 | 1774 | GTEx | DepMap | Descartes | 37.07 | 1373.60 |
| BASP1 | 0.0005485 | 1904 | GTEx | DepMap | Descartes | 20.03 | 53.29 |
| TUBB2B | 0.0004732 | 2193 | GTEx | DepMap | Descartes | 11.20 | 413.41 |
| ISL1 | 0.0003072 | 3137 | GTEx | DepMap | Descartes | 1.70 | 22.60 |
| TUBB2A | 0.0002998 | 3192 | GTEx | DepMap | Descartes | 3.90 | 141.53 |
| EYA1 | 0.0002462 | 3657 | GTEx | DepMap | Descartes | 0.56 | 0.61 |
| RBFOX1 | 0.0002266 | 3848 | GTEx | DepMap | Descartes | 0.34 | 0.03 |
| STMN4 | 0.0001952 | 4170 | GTEx | DepMap | Descartes | 0.61 | 4.73 |
| NTRK1 | 0.0001832 | 4342 | GTEx | DepMap | Descartes | 0.50 | 2.59 |
| MLLT11 | 0.0001825 | 4352 | GTEx | DepMap | Descartes | 1.61 | 14.27 |
| NPY | 0.0001550 | 4737 | GTEx | DepMap | Descartes | 0.90 | 24.07 |
| STMN2 | 0.0001451 | 4891 | GTEx | DepMap | Descartes | 4.88 | 10.33 |
| RPH3A | 0.0001332 | 5101 | GTEx | DepMap | Descartes | 0.02 | 0.05 |
| GAL | 0.0001243 | 5229 | GTEx | DepMap | Descartes | 0.33 | 5.57 |
| MAB21L2 | 0.0001233 | 5256 | GTEx | DepMap | Descartes | 0.66 | 24.11 |
| HS3ST5 | 0.0000815 | 6014 | GTEx | DepMap | Descartes | 0.05 | 0.02 |
| REEP1 | 0.0000645 | 6440 | GTEx | DepMap | Descartes | 0.28 | 0.27 |
| MARCH11 | 0.0000567 | 6640 | GTEx | DepMap | Descartes | 0.09 | 0.11 |
| GAP43 | 0.0000526 | 6747 | GTEx | DepMap | Descartes | 3.99 | 5.10 |
| FAT3 | 0.0000479 | 6854 | GTEx | DepMap | Descartes | 0.34 | 0.09 |
| EPHA6 | 0.0000145 | 7857 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MAP1B | 0.0000098 | 8036 | GTEx | DepMap | Descartes | 7.21 | 9.86 |
| TMEFF2 | 0.0000091 | 8059 | GTEx | DepMap | Descartes | 0.31 | 0.14 |
| KCNB2 | 0.0000054 | 8201 | GTEx | DepMap | Descartes | 0.12 | 0.04 |
| TMEM132C | 0.0000020 | 8326 | GTEx | DepMap | Descartes | 0.56 | 0.30 |
| ELAVL2 | -0.0000012 | 8430 | GTEx | DepMap | Descartes | 1.21 | 1.00 |
| EYA4 | -0.0000220 | 9191 | GTEx | DepMap | Descartes | 0.12 | 0.08 |
| IL7 | -0.0000395 | 9772 | GTEx | DepMap | Descartes | 0.00 | 0.01 |
| ANKFN1 | -0.0000396 | 9781 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
Descartes Vascular_endothelial markers
Top 50 marker genes of Vascular_endothelial cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/vascular_endothelial/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.76e-01
Mean rank of genes in gene set: 11349.67
Median rank of genes in gene set: 12244
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| APLNR | 0.0003978 | 2578 | GTEx | DepMap | Descartes | 0.08 | 3.18 |
| CYP26B1 | 0.0003759 | 2690 | GTEx | DepMap | Descartes | 0.23 | 1.99 |
| PLVAP | 0.0002468 | 3651 | GTEx | DepMap | Descartes | 0.23 | 1.87 |
| ID1 | 0.0002017 | 4099 | GTEx | DepMap | Descartes | 6.41 | 788.24 |
| NR5A2 | 0.0001571 | 4710 | GTEx | DepMap | Descartes | 0.01 | 0.01 |
| ROBO4 | 0.0001329 | 5107 | GTEx | DepMap | Descartes | 0.03 | 0.33 |
| KANK3 | 0.0000658 | 6411 | GTEx | DepMap | Descartes | 0.21 | 2.30 |
| NOTCH4 | 0.0000162 | 7797 | GTEx | DepMap | Descartes | 0.07 | 0.49 |
| KDR | 0.0000151 | 7836 | GTEx | DepMap | Descartes | 0.19 | 0.53 |
| ESM1 | 0.0000082 | 8080 | GTEx | DepMap | Descartes | 0.00 | 0.09 |
| RASIP1 | -0.0000151 | 8941 | GTEx | DepMap | Descartes | 0.12 | 1.26 |
| NPR1 | -0.0000391 | 9765 | GTEx | DepMap | Descartes | 0.05 | 0.45 |
| EHD3 | -0.0000463 | 10021 | GTEx | DepMap | Descartes | 0.27 | 1.38 |
| CDH13 | -0.0000549 | 10340 | GTEx | DepMap | Descartes | 1.12 | 0.12 |
| CRHBP | -0.0000571 | 10411 | GTEx | DepMap | Descartes | 0.00 | 0.02 |
| FLT4 | -0.0000642 | 10636 | GTEx | DepMap | Descartes | 0.03 | 0.08 |
| CHRM3 | -0.0000898 | 11509 | GTEx | DepMap | Descartes | 0.03 | 0.01 |
| PTPRB | -0.0000979 | 11756 | GTEx | DepMap | Descartes | 0.04 | 0.04 |
| FCGR2B | -0.0001057 | 12000 | GTEx | DepMap | Descartes | 0.00 | 0.01 |
| TEK | -0.0001091 | 12087 | GTEx | DepMap | Descartes | 0.09 | 0.10 |
| SHANK3 | -0.0001099 | 12112 | GTEx | DepMap | Descartes | 0.04 | 0.09 |
| SHE | -0.0001142 | 12244 | GTEx | DepMap | Descartes | 0.04 | 0.19 |
| CEACAM1 | -0.0001190 | 12384 | GTEx | DepMap | Descartes | 0.00 | 0.08 |
| TIE1 | -0.0001197 | 12413 | GTEx | DepMap | Descartes | 0.04 | 0.31 |
| SOX18 | -0.0001252 | 12565 | GTEx | DepMap | Descartes | 0.03 | 1.96 |
| GALNT15 | -0.0001354 | 12876 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| BTNL9 | -0.0001401 | 13014 | GTEx | DepMap | Descartes | 0.01 | 0.04 |
| HYAL2 | -0.0001451 | 13135 | GTEx | DepMap | Descartes | 1.96 | 47.31 |
| TMEM88 | -0.0001591 | 13526 | GTEx | DepMap | Descartes | 0.18 | 10.98 |
| CALCRL | -0.0001606 | 13567 | GTEx | DepMap | Descartes | 0.13 | 0.18 |
Descartes stromal markers
Top 50 marker genes of stromal cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/stromal/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 15105.04
Median rank of genes in gene set: 17344
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| FREM1 | 0.0004982 | 2099 | GTEx | DepMap | Descartes | 0.40 | 0.31 |
| GLI2 | 0.0003223 | 3020 | GTEx | DepMap | Descartes | 0.51 | 0.28 |
| PRRX1 | 0.0002117 | 3982 | GTEx | DepMap | Descartes | 2.13 | 5.83 |
| PCDH18 | 0.0001864 | 4291 | GTEx | DepMap | Descartes | 1.62 | 17.34 |
| EDNRA | 0.0001397 | 4972 | GTEx | DepMap | Descartes | 0.48 | 1.18 |
| C7 | 0.0000872 | 5897 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ADAMTSL3 | -0.0000159 | 8973 | GTEx | DepMap | Descartes | 0.03 | 0.02 |
| PDGFRA | -0.0000828 | 11279 | GTEx | DepMap | Descartes | 1.47 | 5.79 |
| LAMC3 | -0.0000918 | 11572 | GTEx | DepMap | Descartes | 0.01 | 0.03 |
| ABCA6 | -0.0001119 | 12175 | GTEx | DepMap | Descartes | 0.02 | 0.04 |
| SULT1E1 | -0.0001285 | 12675 | GTEx | DepMap | Descartes | 0.00 | 0.03 |
| GAS2 | -0.0001317 | 12767 | GTEx | DepMap | Descartes | 1.39 | 1.84 |
| HHIP | -0.0001327 | 12794 | GTEx | DepMap | Descartes | 0.18 | 0.30 |
| FNDC1 | -0.0001491 | 13238 | GTEx | DepMap | Descartes | 0.08 | 0.13 |
| ABCC9 | -0.0001789 | 14047 | GTEx | DepMap | Descartes | 0.13 | 0.13 |
| RSPO3 | -0.0002246 | 14973 | GTEx | DepMap | Descartes | 1.17 | 1.77 |
| ELN | -0.0002497 | 15395 | GTEx | DepMap | Descartes | 0.64 | 2.53 |
| LRRC17 | -0.0003005 | 16147 | GTEx | DepMap | Descartes | 0.97 | 4.69 |
| CLDN11 | -0.0003032 | 16184 | GTEx | DepMap | Descartes | 0.29 | 3.94 |
| BICC1 | -0.0003038 | 16194 | GTEx | DepMap | Descartes | 1.40 | 0.92 |
| PRICKLE1 | -0.0003702 | 16949 | GTEx | DepMap | Descartes | 0.50 | 0.78 |
| CDH11 | -0.0003727 | 16976 | GTEx | DepMap | Descartes | 2.42 | 2.46 |
| ITGA11 | -0.0003734 | 16981 | GTEx | DepMap | Descartes | 0.04 | 0.05 |
| IGFBP3 | -0.0004665 | 17707 | GTEx | DepMap | Descartes | 1.80 | 31.20 |
| MGP | -0.0005423 | 18075 | GTEx | DepMap | Descartes | 2.06 | 91.98 |
| ACTA2 | -0.0005510 | 18102 | GTEx | DepMap | Descartes | 8.80 | 59.30 |
| SCARA5 | -0.0005661 | 18170 | GTEx | DepMap | Descartes | 0.07 | 0.11 |
| ADAMTS2 | -0.0005670 | 18175 | GTEx | DepMap | Descartes | 0.21 | 0.15 |
| PAMR1 | -0.0006653 | 18494 | GTEx | DepMap | Descartes | 0.19 | 0.36 |
| CD248 | -0.0006942 | 18580 | GTEx | DepMap | Descartes | 0.73 | 45.23 |
Descartes sympathoblasts markers
Top 50 marker genes of sympathoblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/sympathoblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 6.04e-01
Mean rank of genes in gene set: 9891.88
Median rank of genes in gene set: 8972
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| HTATSF1 | 0.0007195 | 1422 | GTEx | DepMap | Descartes | 3.35 | 33.08 |
| PCSK1N | 0.0002272 | 3841 | GTEx | DepMap | Descartes | 1.68 | 53.70 |
| TMEM130 | 0.0001912 | 4221 | GTEx | DepMap | Descartes | 0.03 | 0.12 |
| PCSK2 | 0.0001310 | 5135 | GTEx | DepMap | Descartes | 0.25 | 0.09 |
| MGAT4C | 0.0001081 | 5515 | GTEx | DepMap | Descartes | 0.03 | 0.01 |
| INSM1 | 0.0000872 | 5900 | GTEx | DepMap | Descartes | 0.29 | 13.26 |
| TBX20 | 0.0000756 | 6152 | GTEx | DepMap | Descartes | 0.18 | 0.38 |
| FAM155A | 0.0000717 | 6257 | GTEx | DepMap | Descartes | 0.10 | 0.03 |
| SLC18A1 | 0.0000539 | 6717 | GTEx | DepMap | Descartes | 0.10 | 0.30 |
| CHGA | 0.0000500 | 6813 | GTEx | DepMap | Descartes | 0.62 | 10.39 |
| TENM1 | 0.0000406 | 7071 | GTEx | DepMap | Descartes | 0.04 | 0.01 |
| DGKK | 0.0000365 | 7180 | GTEx | DepMap | Descartes | 0.19 | 0.19 |
| KSR2 | 0.0000314 | 7325 | GTEx | DepMap | Descartes | 0.06 | 0.02 |
| CHGB | 0.0000224 | 7612 | GTEx | DepMap | Descartes | 0.98 | 11.01 |
| TIAM1 | 0.0000175 | 7763 | GTEx | DepMap | Descartes | 1.45 | 0.43 |
| CCSER1 | 0.0000162 | 7798 | GTEx | DepMap | Descartes | 0.09 | 0.01 |
| FGF14 | 0.0000042 | 8250 | GTEx | DepMap | Descartes | 0.01 | 0.00 |
| SCG2 | 0.0000022 | 8315 | GTEx | DepMap | Descartes | 0.04 | 1.06 |
| SPOCK3 | -0.0000079 | 8658 | GTEx | DepMap | Descartes | 0.23 | 0.09 |
| ST18 | -0.0000131 | 8874 | GTEx | DepMap | Descartes | 0.08 | 0.03 |
| CDH12 | -0.0000135 | 8887 | GTEx | DepMap | Descartes | 0.02 | 0.00 |
| EML6 | -0.0000185 | 9057 | GTEx | DepMap | Descartes | 0.03 | 0.02 |
| AGBL4 | -0.0000374 | 9703 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ARC | -0.0000410 | 9833 | GTEx | DepMap | Descartes | 0.05 | 1.87 |
| UNC80 | -0.0000466 | 10034 | GTEx | DepMap | Descartes | 0.07 | 0.04 |
| SLC24A2 | -0.0000594 | 10484 | GTEx | DepMap | Descartes | 0.02 | 0.01 |
| CDH18 | -0.0000638 | 10622 | GTEx | DepMap | Descartes | 0.01 | 0.00 |
| KCTD16 | -0.0000911 | 11545 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PNMT | -0.0001070 | 12034 | GTEx | DepMap | Descartes | 0.03 | 2.04 |
| GRM7 | -0.0001248 | 12554 | GTEx | DepMap | Descartes | 0.02 | 0.00 |
Descartes erythroblasts markers
Top 50 marker genes of erythroblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/erythroblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.91e-01
Mean rank of genes in gene set: 11860.36
Median rank of genes in gene set: 12911
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| EPB41 | 0.0004371 | 2358 | GTEx | DepMap | Descartes | 1.84 | 1.47 |
| SPTB | 0.0003234 | 3015 | GTEx | DepMap | Descartes | 0.11 | 0.14 |
| GCLC | 0.0002984 | 3199 | GTEx | DepMap | Descartes | 0.62 | 1.74 |
| TRAK2 | 0.0002163 | 3931 | GTEx | DepMap | Descartes | 0.41 | 0.75 |
| RHAG | 0.0002007 | 4109 | GTEx | DepMap | Descartes | 0.01 | 0.03 |
| XPO7 | 0.0001458 | 4882 | GTEx | DepMap | Descartes | 1.41 | 1.51 |
| TMEM56 | 0.0000908 | 5823 | GTEx | DepMap | Descartes | 0.04 | 0.06 |
| TSPAN5 | 0.0000900 | 5841 | GTEx | DepMap | Descartes | 0.84 | 0.65 |
| ANK1 | 0.0000599 | 6558 | GTEx | DepMap | Descartes | 0.33 | 0.24 |
| SPECC1 | -0.0000203 | 9134 | GTEx | DepMap | Descartes | 0.63 | 0.31 |
| MARCH3 | -0.0000398 | 9785 | GTEx | DepMap | Descartes | 0.03 | 0.03 |
| SLC25A21 | -0.0000553 | 10353 | GTEx | DepMap | Descartes | 0.01 | 0.00 |
| TMCC2 | -0.0000591 | 10471 | GTEx | DepMap | Descartes | 0.27 | 1.01 |
| RAPGEF2 | -0.0000740 | 10968 | GTEx | DepMap | Descartes | 0.65 | 0.38 |
| CPOX | -0.0000824 | 11259 | GTEx | DepMap | Descartes | 0.90 | 2.07 |
| TFR2 | -0.0000917 | 11570 | GTEx | DepMap | Descartes | 0.02 | 0.10 |
| RHD | -0.0001132 | 12214 | GTEx | DepMap | Descartes | 0.04 | 0.13 |
| SPTA1 | -0.0001292 | 12697 | GTEx | DepMap | Descartes | 0.01 | 0.01 |
| DENND4A | -0.0001448 | 13125 | GTEx | DepMap | Descartes | 0.15 | 0.17 |
| GYPC | -0.0001688 | 13789 | GTEx | DepMap | Descartes | 0.38 | 1.73 |
| EPB42 | -0.0001718 | 13864 | GTEx | DepMap | Descartes | 0.00 | 0.01 |
| BLVRB | -0.0001810 | 14095 | GTEx | DepMap | Descartes | 1.13 | 8.66 |
| SNCA | -0.0001935 | 14382 | GTEx | DepMap | Descartes | 0.17 | 0.16 |
| HEMGN | -0.0002073 | 14659 | GTEx | DepMap | Descartes | 0.02 | 0.12 |
| RGS6 | -0.0002513 | 15423 | GTEx | DepMap | Descartes | 0.03 | 0.01 |
| SLC4A1 | -0.0002866 | 15959 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MICAL2 | -0.0003264 | 16489 | GTEx | DepMap | Descartes | 0.15 | 0.09 |
| GYPA | -0.0003325 | 16560 | GTEx | DepMap | Descartes | 0.01 | 0.06 |
| SLC25A37 | -0.0003343 | 16582 | GTEx | DepMap | Descartes | 0.45 | 1.10 |
| CR1L | -0.0003545 | 16789 | GTEx | DepMap | Descartes | 1.13 | 4.22 |
Descartes myeloid markers
Top 50 marker genes of myeloid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/myeloid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 13672.98
Median rank of genes in gene set: 13676.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| WWP1 | 0.0003056 | 3151 | GTEx | DepMap | Descartes | 1.00 | 1.24 |
| HRH1 | 0.0002245 | 3863 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| RBPJ | 0.0001956 | 4162 | GTEx | DepMap | Descartes | 1.75 | 1.21 |
| ADAP2 | 0.0001601 | 4658 | GTEx | DepMap | Descartes | 0.01 | 0.04 |
| CPVL | 0.0001596 | 4667 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MARCH1 | 0.0000712 | 6270 | GTEx | DepMap | Descartes | 0.02 | 0.00 |
| MS4A4A | -0.0000127 | 8859 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CYBB | -0.0000582 | 10456 | GTEx | DepMap | Descartes | 0.04 | 0.07 |
| CSF1R | -0.0000652 | 10672 | GTEx | DepMap | Descartes | 0.05 | 0.20 |
| SLC9A9 | -0.0000744 | 10987 | GTEx | DepMap | Descartes | 0.10 | 0.02 |
| MS4A7 | -0.0000885 | 11470 | GTEx | DepMap | Descartes | 0.00 | 0.02 |
| FMN1 | -0.0001025 | 11900 | GTEx | DepMap | Descartes | 0.18 | 0.05 |
| C1QC | -0.0001211 | 12458 | GTEx | DepMap | Descartes | 0.11 | 4.04 |
| C1QB | -0.0001249 | 12558 | GTEx | DepMap | Descartes | 0.13 | 2.50 |
| RNASE1 | -0.0001270 | 12631 | GTEx | DepMap | Descartes | 0.00 | 0.10 |
| C1QA | -0.0001348 | 12855 | GTEx | DepMap | Descartes | 0.08 | 3.06 |
| SFMBT2 | -0.0001349 | 12859 | GTEx | DepMap | Descartes | 0.07 | 0.04 |
| HCK | -0.0001366 | 12921 | GTEx | DepMap | Descartes | 0.05 | 0.12 |
| CTSS | -0.0001419 | 13061 | GTEx | DepMap | Descartes | 0.05 | 0.19 |
| CD14 | -0.0001439 | 13103 | GTEx | DepMap | Descartes | 0.03 | 1.87 |
| SLCO2B1 | -0.0001569 | 13474 | GTEx | DepMap | Descartes | 0.01 | 0.03 |
| MPEG1 | -0.0001723 | 13879 | GTEx | DepMap | Descartes | 0.02 | 0.39 |
| MSR1 | -0.0002280 | 15028 | GTEx | DepMap | Descartes | 0.00 | 0.01 |
| SPP1 | -0.0002612 | 15586 | GTEx | DepMap | Descartes | 0.03 | 0.55 |
| CTSC | -0.0002619 | 15594 | GTEx | DepMap | Descartes | 0.59 | 2.04 |
| ATP8B4 | -0.0002723 | 15755 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| RGL1 | -0.0002767 | 15824 | GTEx | DepMap | Descartes | 0.55 | 0.32 |
| MERTK | -0.0003588 | 16837 | GTEx | DepMap | Descartes | 0.19 | 0.24 |
| IFNGR1 | -0.0003875 | 17115 | GTEx | DepMap | Descartes | 0.39 | 3.00 |
| LGMN | -0.0004162 | 17348 | GTEx | DepMap | Descartes | 0.91 | 2.39 |
Descartes Schwann markers
Top 50 marker genes of Schwann cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/schwann/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 13974.53
Median rank of genes in gene set: 15082
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| MARCKS | 0.0012617 | 715 | GTEx | DepMap | Descartes | 22.14 | 565.56 |
| HMGA2 | 0.0009282 | 1053 | GTEx | DepMap | Descartes | 15.79 | 18.97 |
| FIGN | 0.0004566 | 2269 | GTEx | DepMap | Descartes | 1.18 | 1.43 |
| NRXN1 | 0.0002721 | 3416 | GTEx | DepMap | Descartes | 0.52 | 0.05 |
| ERBB4 | 0.0001375 | 5013 | GTEx | DepMap | Descartes | 0.07 | 0.01 |
| XKR4 | 0.0000432 | 6989 | GTEx | DepMap | Descartes | 0.04 | NA |
| PAG1 | 0.0000168 | 7781 | GTEx | DepMap | Descartes | 0.39 | 0.34 |
| SCN7A | -0.0000190 | 9075 | GTEx | DepMap | Descartes | 0.03 | 0.03 |
| GRIK3 | -0.0000347 | 9616 | GTEx | DepMap | Descartes | 0.04 | 0.02 |
| NRXN3 | -0.0000536 | 10297 | GTEx | DepMap | Descartes | 0.26 | 0.01 |
| IL1RAPL2 | -0.0000553 | 10352 | GTEx | DepMap | Descartes | 0.02 | 0.00 |
| PPP2R2B | -0.0000646 | 10652 | GTEx | DepMap | Descartes | 0.56 | 0.19 |
| SOX10 | -0.0000797 | 11179 | GTEx | DepMap | Descartes | 0.71 | 11.60 |
| ERBB3 | -0.0000858 | 11377 | GTEx | DepMap | Descartes | 1.16 | 6.05 |
| STARD13 | -0.0001130 | 12209 | GTEx | DepMap | Descartes | 0.33 | 0.23 |
| TRPM3 | -0.0001449 | 13129 | GTEx | DepMap | Descartes | 0.05 | 0.01 |
| LRRTM4 | -0.0001490 | 13236 | GTEx | DepMap | Descartes | 0.02 | 0.00 |
| SLC35F1 | -0.0001568 | 13471 | GTEx | DepMap | Descartes | 0.32 | 0.11 |
| LAMC1 | -0.0001624 | 13616 | GTEx | DepMap | Descartes | 1.58 | 1.57 |
| MDGA2 | -0.0001679 | 13756 | GTEx | DepMap | Descartes | 0.11 | 0.02 |
| SORCS1 | -0.0001824 | 14131 | GTEx | DepMap | Descartes | 0.27 | 0.07 |
| PLCE1 | -0.0001853 | 14189 | GTEx | DepMap | Descartes | 0.48 | 0.24 |
| CDH19 | -0.0001978 | 14456 | GTEx | DepMap | Descartes | 0.17 | 0.26 |
| IL1RAPL1 | -0.0002312 | 15082 | GTEx | DepMap | Descartes | 0.12 | 0.01 |
| GAS7 | -0.0002375 | 15197 | GTEx | DepMap | Descartes | 0.55 | 0.31 |
| PLP1 | -0.0002648 | 15636 | GTEx | DepMap | Descartes | 1.26 | 10.80 |
| SOX5 | -0.0003848 | 17084 | GTEx | DepMap | Descartes | 0.88 | 0.16 |
| PMP22 | -0.0004226 | 17405 | GTEx | DepMap | Descartes | 2.39 | 14.54 |
| FAM134B | -0.0004807 | 17785 | GTEx | DepMap | Descartes | 0.30 | NA |
| DST | -0.0006166 | 18351 | GTEx | DepMap | Descartes | 6.47 | 1.92 |
Descartes Megakaryocytes markers
Top 50 marker genes of Megakaryocytes cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/megakaryocytes/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.38e-01
Mean rank of genes in gene set: 10912.89
Median rank of genes in gene set: 11167
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TPM4 | 0.0013743 | 647 | GTEx | DepMap | Descartes | 6.37 | 35.57 |
| LIMS1 | 0.0013139 | 682 | GTEx | DepMap | Descartes | 2.58 | 3.28 |
| ACTB | 0.0012166 | 750 | GTEx | DepMap | Descartes | 135.53 | 5103.42 |
| ACTN1 | 0.0007455 | 1356 | GTEx | DepMap | Descartes | 2.44 | 3.37 |
| SPN | 0.0006649 | 1553 | GTEx | DepMap | Descartes | 0.01 | 0.14 |
| PRKAR2B | 0.0005349 | 1966 | GTEx | DepMap | Descartes | 1.02 | 1.22 |
| FLI1 | 0.0002916 | 3257 | GTEx | DepMap | Descartes | 0.36 | 0.52 |
| UBASH3B | 0.0001604 | 4654 | GTEx | DepMap | Descartes | 0.28 | 0.30 |
| TLN1 | 0.0001243 | 5230 | GTEx | DepMap | Descartes | 3.79 | 15.79 |
| RAP1B | 0.0001040 | 5601 | GTEx | DepMap | Descartes | 3.60 | 15.45 |
| HIPK2 | 0.0000904 | 5829 | GTEx | DepMap | Descartes | 1.93 | 1.23 |
| STOM | 0.0000771 | 6115 | GTEx | DepMap | Descartes | 0.21 | 1.38 |
| ARHGAP6 | 0.0000549 | 6681 | GTEx | DepMap | Descartes | 0.12 | 0.03 |
| PSTPIP2 | 0.0000443 | 6953 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MMRN1 | 0.0000111 | 7983 | GTEx | DepMap | Descartes | 0.03 | 0.05 |
| ZYX | -0.0000156 | 8957 | GTEx | DepMap | Descartes | 3.73 | 45.76 |
| FERMT3 | -0.0000305 | 9470 | GTEx | DepMap | Descartes | 0.04 | 0.25 |
| STON2 | -0.0000430 | 9906 | GTEx | DepMap | Descartes | 0.29 | 0.18 |
| PDE3A | -0.0000442 | 9946 | GTEx | DepMap | Descartes | 0.33 | 0.18 |
| DOK6 | -0.0000498 | 10159 | GTEx | DepMap | Descartes | 0.01 | 0.00 |
| MED12L | -0.0000499 | 10160 | GTEx | DepMap | Descartes | 0.11 | 0.04 |
| PPBP | -0.0000501 | 10171 | GTEx | DepMap | Descartes | 0.01 | 0.82 |
| MYH9 | -0.0000672 | 10742 | GTEx | DepMap | Descartes | 3.59 | 4.54 |
| TRPC6 | -0.0000795 | 11167 | GTEx | DepMap | Descartes | 0.01 | 0.01 |
| FLNA | -0.0001032 | 11923 | GTEx | DepMap | Descartes | 6.43 | 29.25 |
| MCTP1 | -0.0001106 | 12137 | GTEx | DepMap | Descartes | 0.01 | 0.00 |
| P2RX1 | -0.0001272 | 12636 | GTEx | DepMap | Descartes | 0.00 | 0.03 |
| BIN2 | -0.0001562 | 13447 | GTEx | DepMap | Descartes | 0.01 | 0.03 |
| PF4 | -0.0001952 | 14410 | GTEx | DepMap | Descartes | 0.10 | 16.03 |
| SLC2A3 | -0.0001963 | 14430 | GTEx | DepMap | Descartes | 2.59 | 3.06 |
Descartes Lyphoid markers
Top 50 marker genes of Lyphoid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/lymphoid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 3.03e-01
Mean rank of genes in gene set: 9221.45
Median rank of genes in gene set: 8442
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| MSN | 0.0006343 | 1640 | GTEx | DepMap | Descartes | 1.26 | 2.33 |
| LEF1 | 0.0005666 | 1849 | GTEx | DepMap | Descartes | 2.57 | 2.61 |
| PDE3B | 0.0005587 | 1873 | GTEx | DepMap | Descartes | 0.31 | 0.39 |
| ETS1 | 0.0004512 | 2293 | GTEx | DepMap | Descartes | 0.63 | 0.75 |
| IKZF1 | 0.0004360 | 2363 | GTEx | DepMap | Descartes | 0.02 | 0.02 |
| STK39 | 0.0002759 | 3379 | GTEx | DepMap | Descartes | 0.74 | 0.42 |
| NKG7 | 0.0002719 | 3420 | GTEx | DepMap | Descartes | 0.00 | 0.60 |
| EVL | 0.0002653 | 3475 | GTEx | DepMap | Descartes | 4.39 | 4.58 |
| SP100 | 0.0002287 | 3827 | GTEx | DepMap | Descartes | 0.00 | 0.01 |
| GNG2 | 0.0001577 | 4703 | GTEx | DepMap | Descartes | 1.57 | 2.00 |
| BACH2 | 0.0001459 | 4880 | GTEx | DepMap | Descartes | 0.64 | 0.24 |
| ABLIM1 | 0.0001371 | 5024 | GTEx | DepMap | Descartes | 2.44 | 0.83 |
| SKAP1 | 0.0001281 | 5180 | GTEx | DepMap | Descartes | 0.05 | 0.02 |
| WIPF1 | 0.0001233 | 5257 | GTEx | DepMap | Descartes | 0.35 | 0.53 |
| FYN | 0.0001046 | 5584 | GTEx | DepMap | Descartes | 1.59 | 1.13 |
| FOXP1 | 0.0000937 | 5775 | GTEx | DepMap | Descartes | 6.40 | 1.33 |
| CD44 | 0.0000657 | 6413 | GTEx | DepMap | Descartes | 0.24 | 0.31 |
| TMSB10 | 0.0000628 | 6481 | GTEx | DepMap | Descartes | 62.11 | 5267.11 |
| RCSD1 | 0.0000608 | 6532 | GTEx | DepMap | Descartes | 0.52 | 1.05 |
| FAM65B | 0.0000436 | 6976 | GTEx | DepMap | Descartes | 0.08 | NA |
| ARHGAP15 | 0.0000081 | 8086 | GTEx | DepMap | Descartes | 0.02 | 0.00 |
| RAP1GAP2 | -0.0000112 | 8798 | GTEx | DepMap | Descartes | 0.04 | 0.02 |
| LCP1 | -0.0000113 | 8803 | GTEx | DepMap | Descartes | 0.12 | 0.13 |
| DOCK10 | -0.0000225 | 9205 | GTEx | DepMap | Descartes | 0.20 | 0.12 |
| CELF2 | -0.0000291 | 9428 | GTEx | DepMap | Descartes | 2.95 | 0.39 |
| SCML4 | -0.0000749 | 11008 | GTEx | DepMap | Descartes | 0.01 | 0.02 |
| ARID5B | -0.0000812 | 11218 | GTEx | DepMap | Descartes | 1.73 | 1.17 |
| PTPRC | -0.0000988 | 11783 | GTEx | DepMap | Descartes | 0.01 | 0.01 |
| SORL1 | -0.0001349 | 12861 | GTEx | DepMap | Descartes | 0.52 | 0.37 |
| PRKCH | -0.0001564 | 13458 | GTEx | DepMap | Descartes | 0.47 | 0.25 |
Below shows the ranks on this GEP for immune marker gene lists curated by CellTypist (https://www.celltypist.org/encyclopedia).
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Cycling cells: Cycling monocytes (curated markers)
proliferating monocytes:
Wilcoxon ranksum test P-value for gene set overrepresentation: 4.99e-03
Mean rank of genes in gene set: 1364.67
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TOP2A | 0.0138475 | 12 | GTEx | DepMap | Descartes | 25.38 | 128.60 |
| MKI67 | 0.0133305 | 16 | GTEx | DepMap | Descartes | 13.00 | 72.38 |
| S100A9 | 0.0002043 | 4066 | GTEx | DepMap | Descartes | 0.02 | 0.49 |
Cycling cells: Cycling B cells (curated markers)
proliferating B lymphocytes:
Wilcoxon ranksum test P-value for gene set overrepresentation: 7.22e-03
Mean rank of genes in gene set: 14
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TOP2A | 0.0138475 | 12 | GTEx | DepMap | Descartes | 25.38 | 128.60 |
| MKI67 | 0.0133305 | 16 | GTEx | DepMap | Descartes | 13.00 | 72.38 |
Cycling cells: Cycling gamma-delta T cells (curated markers)
proliferating gamma-delta (γδ) T lymphocytes:
Wilcoxon ranksum test P-value for gene set overrepresentation: 7.22e-03
Mean rank of genes in gene set: 14
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TOP2A | 0.0138475 | 12 | GTEx | DepMap | Descartes | 25.38 | 128.60 |
| MKI67 | 0.0133305 | 16 | GTEx | DepMap | Descartes | 13.00 | 72.38 |