Program: 2. Neuroblastoma #2.
Program description and justification of annotation: 2.
Submit a comment on this gene expression program’s interpretation: CLICK
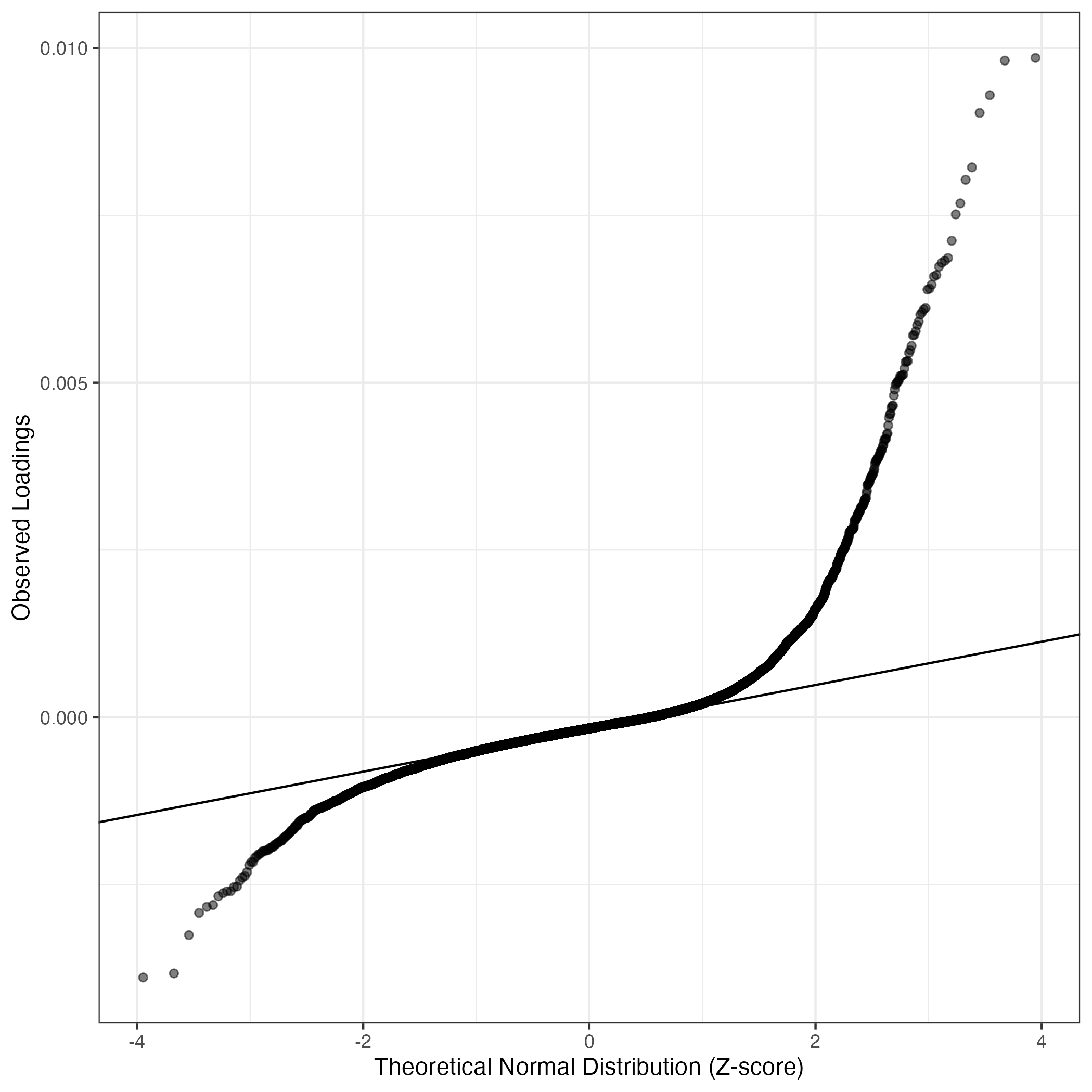
QQ-plot of gene loadings, averaged over both independent splits of the data
This plot highlights the relative contribution of each gene to the GEP
Top genes driving this program.
Note: Decartes website is buggy, try refreshing. Also, Decartes fetal adrenal data have been collected at specific time points (89-122 days), all possible cell types of interest may not be represented, do not overinterpret.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
| Gene | Loading | Gene.Name | GTEx | DepMap | Descartes | Mean.Counts | Mean.Tpm | |
|---|---|---|---|---|---|---|---|---|
| 1 | NRG1 | 0.0098541 | neuregulin 1 | GTEx | DepMap | Descartes | 14.28 | 1676.92 |
| 2 | NXPH1 | 0.0098145 | neurexophilin 1 | GTEx | DepMap | Descartes | 10.69 | 4455.23 |
| 3 | EBF1 | 0.0092964 | EBF transcription factor 1 | GTEx | DepMap | Descartes | 6.50 | 1599.53 |
| 4 | LRRTM4 | 0.0090314 | leucine rich repeat transmembrane neuronal 4 | GTEx | DepMap | Descartes | 6.03 | 2347.41 |
| 5 | RYR2 | 0.0082173 | ryanodine receptor 2 | GTEx | DepMap | Descartes | 7.52 | 588.12 |
| 6 | PTPRD | 0.0080334 | protein tyrosine phosphatase receptor type D | GTEx | DepMap | Descartes | 4.37 | 571.14 |
| 7 | DPP6 | 0.0076801 | dipeptidyl peptidase like 6 | GTEx | DepMap | Descartes | 10.63 | 2854.03 |
| 8 | HS3ST5 | 0.0075159 | heparan sulfate-glucosamine 3-sulfotransferase 5 | GTEx | DepMap | Descartes | 2.70 | 942.71 |
| 9 | CADM1 | 0.0071219 | cell adhesion molecule 1 | GTEx | DepMap | Descartes | 6.18 | 941.18 |
| 10 | RGS7 | 0.0068647 | regulator of G protein signaling 7 | GTEx | DepMap | Descartes | 7.58 | 4037.17 |
| 11 | SDK1 | 0.0068226 | sidekick cell adhesion molecule 1 | GTEx | DepMap | Descartes | 5.61 | 696.61 |
| 12 | BRINP3 | 0.0067927 | BMP/retinoic acid inducible neural specific 3 | GTEx | DepMap | Descartes | 2.27 | NA |
| 13 | OSBPL3 | 0.0067320 | oxysterol binding protein like 3 | GTEx | DepMap | Descartes | 1.90 | 372.34 |
| 14 | ROBO1 | 0.0066089 | roundabout guidance receptor 1 | GTEx | DepMap | Descartes | 5.38 | 976.27 |
| 15 | ROBO2 | 0.0065873 | roundabout guidance receptor 2 | GTEx | DepMap | Descartes | 3.06 | 430.63 |
| 16 | AUTS2 | 0.0064650 | activator of transcription and developmental regulator AUTS2 | GTEx | DepMap | Descartes | 17.39 | 2992.44 |
| 17 | ANKFN1 | 0.0064057 | ankyrin repeat and fibronectin type III domain containing 1 | GTEx | DepMap | Descartes | 1.36 | 440.69 |
| 18 | DPYD | 0.0063915 | dihydropyrimidine dehydrogenase | GTEx | DepMap | Descartes | 2.66 | 811.70 |
| 19 | ELAVL2 | 0.0061155 | ELAV like RNA binding protein 2 | GTEx | DepMap | Descartes | 2.84 | 961.56 |
| 20 | DCC | 0.0060933 | DCC netrin 1 receptor | GTEx | DepMap | Descartes | 1.78 | 238.65 |
| 21 | SORBS2 | 0.0060529 | sorbin and SH3 domain containing 2 | GTEx | DepMap | Descartes | 4.99 | 1088.21 |
| 22 | SOX6 | 0.0060181 | SRY-box transcription factor 6 | GTEx | DepMap | Descartes | 1.65 | 246.18 |
| 23 | CTNND2 | 0.0059146 | catenin delta 2 | GTEx | DepMap | Descartes | 3.37 | 764.51 |
| 24 | DLC1 | 0.0058602 | DLC1 Rho GTPase activating protein | GTEx | DepMap | Descartes | 8.85 | 1568.03 |
| 25 | AGBL4 | 0.0057700 | AGBL carboxypeptidase 4 | GTEx | DepMap | Descartes | 2.85 | 941.23 |
| 26 | ITGA8 | 0.0057133 | integrin subunit alpha 8 | GTEx | DepMap | Descartes | 1.06 | 221.14 |
| 27 | CACNA2D3 | 0.0057060 | calcium voltage-gated channel auxiliary subunit alpha2delta 3 | GTEx | DepMap | Descartes | 5.76 | 2038.29 |
| 28 | ALCAM | 0.0055512 | activated leukocyte cell adhesion molecule | GTEx | DepMap | Descartes | 2.86 | 840.39 |
| 29 | DGKB | 0.0054850 | diacylglycerol kinase beta | GTEx | DepMap | Descartes | 5.23 | 1046.73 |
| 30 | MALAT1 | 0.0054443 | metastasis associated lung adenocarcinoma transcript 1 | GTEx | DepMap | Descartes | 307.08 | 46779.88 |
| 31 | IL7 | 0.0053210 | interleukin 7 | GTEx | DepMap | Descartes | 3.06 | 2006.66 |
| 32 | EPB41L4A | 0.0053194 | erythrocyte membrane protein band 4.1 like 4A | GTEx | DepMap | Descartes | 0.92 | 264.24 |
| 33 | SYN3 | 0.0053095 | synapsin III | GTEx | DepMap | Descartes | 3.58 | 593.85 |
| 34 | ZBTB20 | 0.0052120 | zinc finger and BTB domain containing 20 | GTEx | DepMap | Descartes | 3.46 | 171.75 |
| 35 | SEMA3E | 0.0051211 | semaphorin 3E | GTEx | DepMap | Descartes | 1.13 | 209.98 |
| 36 | LRRC4C | 0.0051119 | leucine rich repeat containing 4C | GTEx | DepMap | Descartes | 1.75 | 576.25 |
| 37 | GRIA1 | 0.0051029 | glutamate ionotropic receptor AMPA type subunit 1 | GTEx | DepMap | Descartes | 1.12 | 261.71 |
| 38 | SLC30A8 | 0.0050996 | solute carrier family 30 member 8 | GTEx | DepMap | Descartes | 0.59 | 130.77 |
| 39 | RAPGEF5 | 0.0050478 | Rap guanine nucleotide exchange factor 5 | GTEx | DepMap | Descartes | 0.95 | 189.92 |
| 40 | CNTNAP2 | 0.0050173 | contactin associated protein 2 | GTEx | DepMap | Descartes | 16.58 | 2213.78 |
| 41 | KCND2 | 0.0050161 | potassium voltage-gated channel subfamily D member 2 | GTEx | DepMap | Descartes | 6.19 | 1396.67 |
| 42 | UNC5D | 0.0049917 | unc-5 netrin receptor D | GTEx | DepMap | Descartes | 1.50 | 222.38 |
| 43 | FSTL5 | 0.0049685 | follistatin like 5 | GTEx | DepMap | Descartes | 0.91 | 260.51 |
| 44 | LINGO2 | 0.0048978 | leucine rich repeat and Ig domain containing 2 | GTEx | DepMap | Descartes | 5.89 | 2598.64 |
| 45 | FAM155A | 0.0048086 | NA | GTEx | DepMap | Descartes | 9.53 | 1365.94 |
| 46 | DNM3 | 0.0046580 | dynamin 3 | GTEx | DepMap | Descartes | 2.48 | 433.43 |
| 47 | MEI4 | 0.0046544 | meiotic double-stranded break formation protein 4 | GTEx | DepMap | Descartes | 0.76 | NA |
| 48 | GRID2 | 0.0046215 | glutamate ionotropic receptor delta type subunit 2 | GTEx | DepMap | Descartes | 1.54 | 373.20 |
| 49 | TENM3 | 0.0045382 | teneurin transmembrane protein 3 | GTEx | DepMap | Descartes | 5.35 | NA |
| 50 | B3GALT1 | 0.0045265 | beta-1,3-galactosyltransferase 1 | GTEx | DepMap | Descartes | 1.08 | 282.29 |
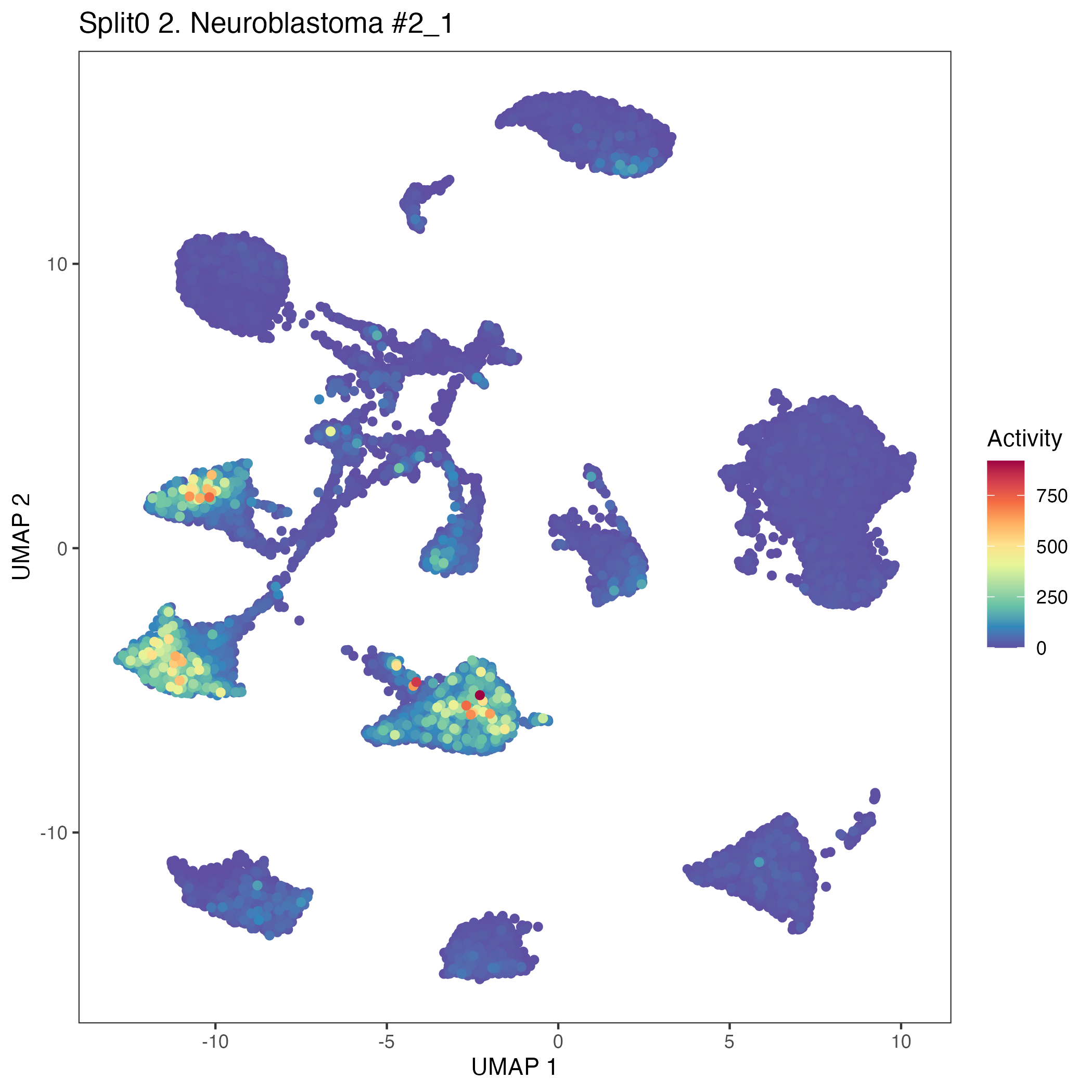
UMAP plots showing activity of gene expression program identified in community:2. Neuroblastoma #2
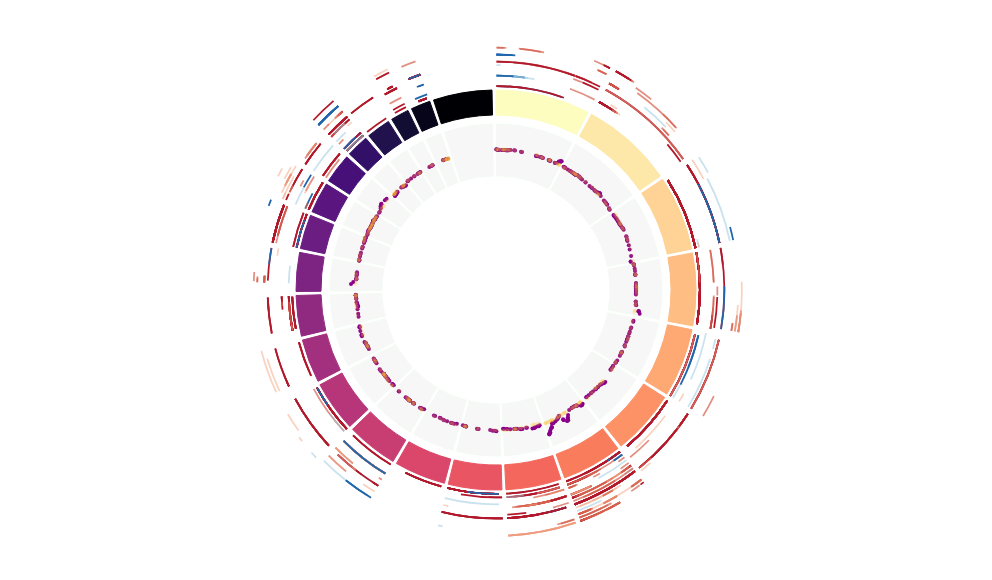
CNV Data procured from inferCNV.
Outer tracks are putative CNV regions (gains = red, losses = blue) for each patient
Inner track is expression data representing:
The top cells expressing this GEP (purple)
Random cells (n =50) from the reference set used in inferCNV (orange)
Gene set Enrichments for this program, caculated from top 50 genes
mSigDB Cell Types Gene Set:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| LAKE_ADULT_KIDNEY_C29_UNKNOWN_NOVEL_PT_CFH_POS_SUBPOPULATION_S2 | 1.04e-06 | 21.30 | 7.21 | 9.56e-05 | 6.99e-04 | 6AUTS2, DPYD, AGBL4, ZBTB20, FAM155A, TENM3 |
86 |
| ZHONG_PFC_MAJOR_TYPES_OPC | 1.45e-06 | 20.04 | 6.80 | 1.08e-04 | 9.74e-04 | 6NXPH1, BRINP3, SOX6, ALCAM, LRRC4C, GRID2 |
91 |
| DESCARTES_FETAL_MUSCLE_SCHWANN_CELLS | 6.23e-06 | 15.35 | 5.24 | 2.99e-04 | 4.18e-03 | 6LRRTM4, CADM1, ALCAM, FSTL5, GRID2, TENM3 |
117 |
| LAKE_ADULT_KIDNEY_C27_VASCULAR_SMOOTH_MUSCLE_CELLS_AND_PERICYTES | 2.26e-06 | 13.75 | 5.13 | 1.52e-04 | 1.52e-03 | 7EBF1, SDK1, AUTS2, SORBS2, DLC1, DGKB, GRID2 |
155 |
| DESCARTES_MAIN_FETAL_OLIGODENDROCYTES | 7.93e-05 | 20.12 | 5.08 | 3.13e-03 | 5.32e-02 | 4BRINP3, SOX6, LRRC4C, KCND2 |
58 |
| DESCARTES_FETAL_THYMUS_THYMIC_EPITHELIAL_CELLS | 3.78e-07 | 11.63 | 4.91 | 4.66e-05 | 2.54e-04 | 9DPP6, SDK1, ROBO2, ANKFN1, SORBS2, EPB41L4A, LRRC4C, UNC5D, TENM3 |
245 |
| MANNO_MIDBRAIN_NEUROTYPES_HOPC | 1.13e-07 | 9.93 | 4.55 | 2.53e-05 | 7.58e-05 | 11NXPH1, LRRTM4, DPP6, BRINP3, SOX6, CTNND2, LRRC4C, KCND2, FAM155A, DNM3, GRID2 |
366 |
| LAKE_ADULT_KIDNEY_C4_PROXIMAL_TUBULE_EPITHELIAL_CELLS_S2 | 5.20e-06 | 12.04 | 4.50 | 2.78e-04 | 3.49e-03 | 7PTPRD, SDK1, SORBS2, SOX6, AGBL4, EPB41L4A, ZBTB20 |
176 |
| LAKE_ADULT_KIDNEY_C26_MESANGIAL_CELLS | 5.39e-06 | 11.97 | 4.48 | 2.78e-04 | 3.62e-03 | 7EBF1, PTPRD, ROBO1, DPYD, DLC1, ITGA8, RAPGEF5 |
177 |
| MANNO_MIDBRAIN_NEUROTYPES_HNBGABA | 3.01e-09 | 8.57 | 4.39 | 1.01e-06 | 2.02e-06 | 16LRRTM4, RYR2, DPP6, CADM1, BRINP3, ROBO1, ROBO2, ELAVL2, SYN3, SEMA3E, GRIA1, CNTNAP2, KCND2, FAM155A, DNM3, GRID2 |
703 |
| MANNO_MIDBRAIN_NEUROTYPES_HNBML1 | 4.17e-07 | 9.83 | 4.34 | 4.66e-05 | 2.80e-04 | 10EBF1, LRRTM4, ROBO1, ROBO2, ELAVL2, DCC, SYN3, SEMA3E, RAPGEF5, FAM155A |
328 |
| MANNO_MIDBRAIN_NEUROTYPES_HOMTN | 2.05e-07 | 9.33 | 4.27 | 3.44e-05 | 1.38e-04 | 11CADM1, ELAVL2, ALCAM, SYN3, SEMA3E, RAPGEF5, CNTNAP2, FSTL5, FAM155A, DNM3, TENM3 |
389 |
| MANNO_MIDBRAIN_NEUROTYPES_HGABA | 5.51e-10 | 7.69 | 4.12 | 3.70e-07 | 3.70e-07 | 20NXPH1, EBF1, RYR2, DPP6, CADM1, ROBO1, ROBO2, AUTS2, ELAVL2, DCC, CTNND2, ITGA8, SYN3, SEMA3E, GRIA1, RAPGEF5, CNTNAP2, FAM155A, DNM3, GRID2 |
1105 |
| DESCARTES_FETAL_ADRENAL_CHROMAFFIN_CELLS | 1.02e-04 | 12.18 | 3.70 | 3.79e-03 | 6.82e-02 | 5RYR2, HS3ST5, ANKFN1, ELAVL2, IL7 |
119 |
| MANNO_MIDBRAIN_NEUROTYPES_HNBML5 | 1.14e-06 | 7.77 | 3.56 | 9.56e-05 | 7.65e-04 | 11RYR2, ROBO1, ROBO2, AUTS2, ELAVL2, DCC, GRIA1, RAPGEF5, CNTNAP2, FAM155A, DNM3 |
465 |
| MANNO_MIDBRAIN_NEUROTYPES_HDA | 2.52e-06 | 7.13 | 3.27 | 1.54e-04 | 1.69e-03 | 11EBF1, DPP6, CADM1, ROBO1, ROBO2, ELAVL2, SEMA3E, GRIA1, RAPGEF5, FAM155A, DNM3 |
506 |
| LAKE_ADULT_KIDNEY_C22_ENDOTHELIAL_CELLS_GLOMERULAR_CAPILLARIES | 1.96e-04 | 10.52 | 3.21 | 6.57e-03 | 1.31e-01 | 5EBF1, DPYD, SORBS2, ITGA8, EPB41L4A |
137 |
| FAN_EMBRYONIC_CTX_OPC | 1.33e-03 | 15.33 | 2.96 | 3.57e-02 | 8.94e-01 | 3NXPH1, BRINP3, LRRC4C |
55 |
| MANNO_MIDBRAIN_NEUROTYPES_HDA1 | 9.39e-06 | 6.16 | 2.83 | 4.20e-04 | 6.30e-03 | 11LRRTM4, DPP6, CADM1, ROBO1, ROBO2, ELAVL2, DCC, RAPGEF5, KCND2, FAM155A, DNM3 |
584 |
| MANNO_MIDBRAIN_NEUROTYPES_HDA2 | 1.99e-05 | 6.22 | 2.76 | 8.36e-04 | 1.34e-02 | 10LRRTM4, DPP6, ROBO1, ROBO2, ELAVL2, DCC, SOX6, CNTNAP2, FAM155A, DNM3 |
513 |
Dowload full table
mSigDB Hallmark Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| HALLMARK_KRAS_SIGNALING_DN | 4.35e-02 | 4.05 | 0.80 | 1.00e+00 | 1.00e+00 | 3RYR2, DCC, GRID2 |
200 |
| HALLMARK_MITOTIC_SPINDLE | 1.83e-01 | 2.64 | 0.31 | 1.00e+00 | 1.00e+00 | 2SORBS2, RAPGEF5 |
199 |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | 1.85e-01 | 2.63 | 0.31 | 1.00e+00 | 1.00e+00 | 2AUTS2, IL7 |
200 |
| HALLMARK_CHOLESTEROL_HOMEOSTASIS | 2.55e-01 | 3.49 | 0.09 | 1.00e+00 | 1.00e+00 | 1ALCAM |
74 |
| HALLMARK_IL6_JAK_STAT3_SIGNALING | 2.93e-01 | 2.97 | 0.07 | 1.00e+00 | 1.00e+00 | 1IL7 |
87 |
| HALLMARK_INTERFERON_ALPHA_RESPONSE | 3.20e-01 | 2.66 | 0.07 | 1.00e+00 | 1.00e+00 | 1IL7 |
97 |
| HALLMARK_PEROXISOME | 3.39e-01 | 2.48 | 0.06 | 1.00e+00 | 1.00e+00 | 1CADM1 |
104 |
| HALLMARK_SPERMATOGENESIS | 4.15e-01 | 1.90 | 0.05 | 1.00e+00 | 1.00e+00 | 1DCC |
135 |
| HALLMARK_UV_RESPONSE_DN | 4.35e-01 | 1.78 | 0.04 | 1.00e+00 | 1.00e+00 | 1DLC1 |
144 |
| HALLMARK_UV_RESPONSE_UP | 4.66e-01 | 1.62 | 0.04 | 1.00e+00 | 1.00e+00 | 1PTPRD |
158 |
| HALLMARK_APOPTOSIS | 4.72e-01 | 1.59 | 0.04 | 1.00e+00 | 1.00e+00 | 1DPYD |
161 |
| HALLMARK_IL2_STAT5_SIGNALING | 5.45e-01 | 1.29 | 0.03 | 1.00e+00 | 1.00e+00 | 1ALCAM |
199 |
| HALLMARK_ESTROGEN_RESPONSE_EARLY | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1DLC1 |
200 |
| HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1CADM1 |
200 |
| HALLMARK_ALLOGRAFT_REJECTION | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1IL7 |
200 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
200 |
| HALLMARK_HYPOXIA | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
200 |
| HALLMARK_WNT_BETA_CATENIN_SIGNALING | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
42 |
| HALLMARK_TGF_BETA_SIGNALING | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
54 |
| HALLMARK_DNA_REPAIR | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
150 |
Dowload full table
KEGG Pathways:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| KEGG_AXON_GUIDANCE | 1.09e-05 | 13.86 | 4.74 | 2.02e-03 | 2.02e-03 | 6ROBO1, ROBO2, DCC, SEMA3E, LRRC4C, UNC5D |
129 |
| KEGG_ARRHYTHMOGENIC_RIGHT_VENTRICULAR_CARDIOMYOPATHY_ARVC | 3.11e-03 | 11.23 | 2.19 | 1.93e-01 | 5.79e-01 | 3RYR2, ITGA8, CACNA2D3 |
74 |
| KEGG_CELL_ADHESION_MOLECULES_CAMS | 1.83e-03 | 8.43 | 2.17 | 1.70e-01 | 3.41e-01 | 4CADM1, ITGA8, ALCAM, CNTNAP2 |
133 |
| KEGG_HYPERTROPHIC_CARDIOMYOPATHY_HCM | 4.29e-03 | 9.97 | 1.94 | 2.00e-01 | 7.98e-01 | 3RYR2, ITGA8, CACNA2D3 |
83 |
| KEGG_DILATED_CARDIOMYOPATHY | 5.37e-03 | 9.17 | 1.79 | 2.00e-01 | 1.00e+00 | 3RYR2, ITGA8, CACNA2D3 |
90 |
| KEGG_LONG_TERM_DEPRESSION | 3.13e-02 | 7.66 | 0.88 | 9.71e-01 | 1.00e+00 | 2GRIA1, GRID2 |
70 |
| KEGG_CARDIAC_MUSCLE_CONTRACTION | 3.90e-02 | 6.76 | 0.78 | 1.00e+00 | 1.00e+00 | 2RYR2, CACNA2D3 |
79 |
| KEGG_PANTOTHENATE_AND_COA_BIOSYNTHESIS | 6.18e-02 | 16.98 | 0.40 | 1.00e+00 | 1.00e+00 | 1DPYD |
16 |
| KEGG_BETA_ALANINE_METABOLISM | 8.40e-02 | 12.14 | 0.29 | 1.00e+00 | 1.00e+00 | 1DPYD |
22 |
| KEGG_GLYCOSPHINGOLIPID_BIOSYNTHESIS_LACTO_AND_NEOLACTO_SERIES | 9.85e-02 | 10.19 | 0.24 | 1.00e+00 | 1.00e+00 | 1B3GALT1 |
26 |
| KEGG_GLYCOSAMINOGLYCAN_BIOSYNTHESIS_HEPARAN_SULFATE | 9.85e-02 | 10.19 | 0.24 | 1.00e+00 | 1.00e+00 | 1HS3ST5 |
26 |
| KEGG_NEUROACTIVE_LIGAND_RECEPTOR_INTERACTION | 2.87e-01 | 1.93 | 0.23 | 1.00e+00 | 1.00e+00 | 2GRIA1, GRID2 |
272 |
| KEGG_GLYCEROLIPID_METABOLISM | 1.77e-01 | 5.31 | 0.13 | 1.00e+00 | 1.00e+00 | 1DGKB |
49 |
| KEGG_DRUG_METABOLISM_OTHER_ENZYMES | 1.84e-01 | 5.10 | 0.12 | 1.00e+00 | 1.00e+00 | 1DPYD |
51 |
| KEGG_AMYOTROPHIC_LATERAL_SCLEROSIS_ALS | 1.90e-01 | 4.91 | 0.12 | 1.00e+00 | 1.00e+00 | 1GRIA1 |
53 |
| KEGG_COLORECTAL_CANCER | 2.19e-01 | 4.18 | 0.10 | 1.00e+00 | 1.00e+00 | 1DCC |
62 |
| KEGG_LONG_TERM_POTENTIATION | 2.43e-01 | 3.70 | 0.09 | 1.00e+00 | 1.00e+00 | 1GRIA1 |
70 |
| KEGG_PHOSPHATIDYLINOSITOL_SIGNALING_SYSTEM | 2.61e-01 | 3.40 | 0.08 | 1.00e+00 | 1.00e+00 | 1DGKB |
76 |
| KEGG_GLYCEROPHOSPHOLIPID_METABOLISM | 2.64e-01 | 3.36 | 0.08 | 1.00e+00 | 1.00e+00 | 1DGKB |
77 |
| KEGG_ECM_RECEPTOR_INTERACTION | 2.84e-01 | 3.07 | 0.08 | 1.00e+00 | 1.00e+00 | 1ITGA8 |
84 |
Dowload full table
CHR Positional Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| chr1q43 | 2.36e-02 | 8.98 | 1.03 | 1.00e+00 | 1.00e+00 | 2RYR2, RGS7 |
60 |
| chr3p12 | 2.81e-02 | 8.14 | 0.94 | 1.00e+00 | 1.00e+00 | 2ROBO1, ROBO2 |
66 |
| chr8p12 | 2.89e-02 | 8.01 | 0.92 | 1.00e+00 | 1.00e+00 | 2NRG1, UNC5D |
67 |
| chr7p21 | 4.26e-02 | 6.43 | 0.74 | 1.00e+00 | 1.00e+00 | 2NXPH1, DGKB |
83 |
| chr7p15 | 5.52e-02 | 5.54 | 0.64 | 1.00e+00 | 1.00e+00 | 2OSBPL3, RAPGEF5 |
96 |
| chr5q33 | 6.89e-02 | 4.87 | 0.57 | 1.00e+00 | 1.00e+00 | 2EBF1, GRIA1 |
109 |
| chr11p12 | 7.30e-02 | 14.16 | 0.33 | 1.00e+00 | 1.00e+00 | 1LRRC4C |
19 |
| chr9p23 | 7.30e-02 | 14.16 | 0.33 | 1.00e+00 | 1.00e+00 | 1PTPRD |
19 |
| chr3q13 | 1.67e-01 | 2.82 | 0.33 | 1.00e+00 | 1.00e+00 | 2ALCAM, ZBTB20 |
187 |
| chr8p22 | 1.64e-01 | 5.80 | 0.14 | 1.00e+00 | 1.00e+00 | 1DLC1 |
45 |
| chr2p12 | 1.90e-01 | 4.91 | 0.12 | 1.00e+00 | 1.00e+00 | 1LRRTM4 |
53 |
| chr13q33 | 1.97e-01 | 4.72 | 0.12 | 1.00e+00 | 1.00e+00 | 1FAM155A |
55 |
| chr5q22 | 1.97e-01 | 4.72 | 0.12 | 1.00e+00 | 1.00e+00 | 1EPB41L4A |
55 |
| chr1p33 | 2.13e-01 | 4.32 | 0.11 | 1.00e+00 | 1.00e+00 | 1AGBL4 |
60 |
| chr10p13 | 2.25e-01 | 4.05 | 0.10 | 1.00e+00 | 1.00e+00 | 1ITGA8 |
64 |
| chr7q35 | 2.28e-01 | 3.99 | 0.10 | 1.00e+00 | 1.00e+00 | 1CNTNAP2 |
65 |
| chr4q22 | 2.43e-01 | 3.70 | 0.09 | 1.00e+00 | 1.00e+00 | 1GRID2 |
70 |
| chr1q31 | 2.46e-01 | 3.64 | 0.09 | 1.00e+00 | 1.00e+00 | 1BRINP3 |
71 |
| chr17q22 | 2.90e-01 | 3.00 | 0.07 | 1.00e+00 | 1.00e+00 | 1ANKFN1 |
86 |
| chr6q14 | 3.09e-01 | 2.77 | 0.07 | 1.00e+00 | 1.00e+00 | 1MEI4 |
93 |
Dowload full table
Transcription Factor Targets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| EN1_01 | 9.45e-04 | 10.15 | 2.61 | 1.99e-01 | 1.00e+00 | 4NRG1, SORBS2, GRIA1, RAPGEF5 |
111 |
| OCT1_Q6 | 6.26e-04 | 6.41 | 2.21 | 1.99e-01 | 7.10e-01 | 6NXPH1, EBF1, CADM1, BRINP3, DLC1, ZBTB20 |
272 |
| STAT5A_04 | 1.45e-03 | 6.64 | 2.04 | 1.99e-01 | 1.00e+00 | 5PTPRD, ELAVL2, AGBL4, GRIA1, GRID2 |
214 |
| SOX9_B1 | 2.29e-03 | 5.96 | 1.83 | 1.99e-01 | 1.00e+00 | 5EBF1, CADM1, ELAVL2, SORBS2, DLC1 |
238 |
| TCCATTKW_UNKNOWN | 2.42e-03 | 5.88 | 1.81 | 1.99e-01 | 1.00e+00 | 5LRRTM4, ELAVL2, SORBS2, CTNND2, CACNA2D3 |
241 |
| TAATTA_CHX10_01 | 7.99e-04 | 3.85 | 1.71 | 1.99e-01 | 9.05e-01 | 10NXPH1, CADM1, ROBO1, ELAVL2, SORBS2, CTNND2, DLC1, ITGA8, CACNA2D3, ZBTB20 |
823 |
| T3R_Q6 | 3.12e-03 | 5.53 | 1.70 | 1.99e-01 | 1.00e+00 | 5LRRTM4, RGS7, ELAVL2, CTNND2, CACNA2D3 |
256 |
| TCF11_01 | 3.23e-03 | 5.49 | 1.69 | 1.99e-01 | 1.00e+00 | 5NRG1, EBF1, LRRTM4, CADM1, GRIA1 |
258 |
| SRY_02 | 3.28e-03 | 5.47 | 1.68 | 1.99e-01 | 1.00e+00 | 5EBF1, CADM1, ELAVL2, SOX6, RAPGEF5 |
259 |
| CDX2_Q5 | 3.50e-03 | 5.38 | 1.65 | 1.99e-01 | 1.00e+00 | 5CADM1, ROBO1, DPYD, ELAVL2, AGBL4 |
263 |
| OCT_Q6 | 3.73e-03 | 5.30 | 1.63 | 1.99e-01 | 1.00e+00 | 5NXPH1, EBF1, CADM1, BRINP3, FSTL5 |
267 |
| AFP1_Q6 | 3.79e-03 | 5.28 | 1.62 | 1.99e-01 | 1.00e+00 | 5HS3ST5, DPYD, ELAVL2, CACNA2D3, CNTNAP2 |
268 |
| OCT1_B | 3.84e-03 | 5.26 | 1.62 | 1.99e-01 | 1.00e+00 | 5NXPH1, EBF1, LRRTM4, CADM1, FSTL5 |
269 |
| OCT_C | 3.90e-03 | 5.24 | 1.61 | 1.99e-01 | 1.00e+00 | 5EBF1, CADM1, ZBTB20, FSTL5, GRID2 |
270 |
| STAT6_01 | 4.03e-03 | 5.20 | 1.60 | 1.99e-01 | 1.00e+00 | 5RYR2, CADM1, BRINP3, CACNA2D3, GRID2 |
272 |
| CRX_Q4 | 4.15e-03 | 5.16 | 1.59 | 1.99e-01 | 1.00e+00 | 5LRRTM4, DPYD, DLC1, ZBTB20, GRID2 |
274 |
| AACTTT_UNKNOWN | 5.19e-04 | 3.08 | 1.58 | 1.99e-01 | 5.88e-01 | 16NRG1, NXPH1, EBF1, LRRTM4, CADM1, BRINP3, AUTS2, ELAVL2, SOX6, CTNND2, CACNA2D3, ALCAM, ZBTB20, RAPGEF5, CNTNAP2, GRID2 |
1928 |
| GR_Q6 | 4.21e-03 | 5.14 | 1.58 | 1.99e-01 | 1.00e+00 | 5BRINP3, AUTS2, ELAVL2, CTNND2, KCND2 |
275 |
| OCT1_Q5_01 | 4.21e-03 | 5.14 | 1.58 | 1.99e-01 | 1.00e+00 | 5NXPH1, EBF1, CADM1, BRINP3, FSTL5 |
275 |
| CDP_02 | 9.53e-03 | 7.39 | 1.45 | 3.18e-01 | 1.00e+00 | 3CADM1, CACNA2D3, DGKB |
111 |
Dowload full table
GO Biological Processes:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GOBP_AXON_MIDLINE_CHOICE_POINT_RECOGNITION | 1.54e-04 | 172.68 | 14.09 | 6.07e-02 | 1.00e+00 | 2ROBO1, ROBO2 |
5 |
| GOBP_OLFACTORY_BULB_INTERNEURON_DEVELOPMENT | 2.31e-04 | 129.66 | 11.45 | 7.53e-02 | 1.00e+00 | 2ROBO1, ROBO2 |
6 |
| GOBP_HETEROPHILIC_CELL_CELL_ADHESION_VIA_PLASMA_MEMBRANE_CELL_ADHESION_MOLECULES | 1.09e-06 | 33.01 | 9.74 | 2.79e-03 | 8.13e-03 | 5PTPRD, CADM1, ALCAM, GRID2, TENM3 |
47 |
| GOBP_AXON_CHOICE_POINT_RECOGNITION | 3.22e-04 | 103.49 | 9.64 | 9.27e-02 | 1.00e+00 | 2ROBO1, ROBO2 |
7 |
| GOBP_NEGATIVE_CHEMOTAXIS | 3.17e-05 | 25.85 | 6.47 | 1.54e-02 | 2.37e-01 | 4NRG1, ROBO1, ROBO2, SEMA3E |
46 |
| GOBP_OLFACTORY_BULB_INTERNEURON_DIFFERENTIATION | 8.35e-04 | 57.69 | 5.91 | 1.69e-01 | 1.00e+00 | 2ROBO1, ROBO2 |
11 |
| GOBP_REGULATION_OF_POSTSYNAPTIC_SPECIALIZATION_ASSEMBLY | 8.35e-04 | 57.69 | 5.91 | 1.69e-01 | 1.00e+00 | 2PTPRD, GRID2 |
11 |
| GOBP_REGULATION_OF_HEART_MORPHOGENESIS | 8.35e-04 | 57.69 | 5.91 | 1.69e-01 | 1.00e+00 | 2ROBO1, ROBO2 |
11 |
| GOBP_CELL_CELL_ADHESION_VIA_PLASMA_MEMBRANE_ADHESION_MOLECULES | 7.87e-08 | 11.88 | 5.24 | 5.89e-04 | 5.89e-04 | 10PTPRD, CADM1, SDK1, ROBO1, ROBO2, ALCAM, LRRC4C, UNC5D, GRID2, TENM3 |
273 |
| GOBP_NETRIN_ACTIVATED_SIGNALING_PATHWAY | 1.18e-03 | 47.19 | 4.96 | 2.00e-01 | 1.00e+00 | 2DCC, UNC5D |
13 |
| GOBP_REGULATION_OF_EXCITATORY_SYNAPSE_ASSEMBLY | 1.18e-03 | 47.19 | 4.96 | 2.00e-01 | 1.00e+00 | 2PTPRD, GRID2 |
13 |
| GOBP_REGULATION_OF_POSTSYNAPTIC_DENSITY_ORGANIZATION | 1.37e-03 | 43.30 | 4.59 | 2.28e-01 | 1.00e+00 | 2PTPRD, GRID2 |
14 |
| GOBP_PULMONARY_VALVE_MORPHOGENESIS | 2.03e-03 | 34.68 | 3.75 | 3.08e-01 | 1.00e+00 | 2ROBO1, ROBO2 |
17 |
| GOBP_CENTRAL_NERVOUS_SYSTEM_NEURON_DEVELOPMENT | 2.89e-04 | 14.11 | 3.60 | 9.01e-02 | 1.00e+00 | 4ROBO1, ROBO2, DCC, AGBL4 |
81 |
| GOBP_NEURON_RECOGNITION | 7.92e-04 | 18.54 | 3.56 | 1.69e-01 | 1.00e+00 | 3ROBO1, ROBO2, CNTNAP2 |
46 |
| GOBP_HEART_FIELD_SPECIFICATION | 2.28e-03 | 32.49 | 3.53 | 3.28e-01 | 1.00e+00 | 2ROBO1, ROBO2 |
18 |
| GOBP_SYNAPSE_ASSEMBLY | 5.61e-05 | 10.21 | 3.51 | 2.47e-02 | 4.20e-01 | 6PTPRD, SDK1, ROBO2, LINGO2, DNM3, GRID2 |
173 |
| GOBP_SYNAPSE_ORGANIZATION | 3.11e-06 | 7.78 | 3.44 | 4.65e-03 | 2.33e-02 | 10PTPRD, SDK1, ROBO2, CTNND2, DGKB, SEMA3E, LRRC4C, LINGO2, DNM3, GRID2 |
412 |
| GOBP_REGULATION_OF_POSTSYNAPSE_ORGANIZATION | 3.79e-04 | 13.08 | 3.35 | 1.05e-01 | 1.00e+00 | 4PTPRD, DGKB, DNM3, GRID2 |
87 |
| GOBP_RETINAL_GANGLION_CELL_AXON_GUIDANCE | 2.54e-03 | 30.58 | 3.34 | 3.52e-01 | 1.00e+00 | 2ROBO2, ALCAM |
19 |
Dowload full table
Immunological Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GSE6259_FLT3L_INDUCED_33D1_POS_DC_VS_CD8_TCELL_DN | 2.83e-03 | 7.44 | 1.92 | 1.00e+00 | 1.00e+00 | 4CADM1, OSBPL3, ITGA8, ALCAM |
150 |
| GSE37605_TREG_VS_TCONV_C57BL6_FOXP3_FUSION_GFP_DN | 5.06e-03 | 6.28 | 1.62 | 1.00e+00 | 1.00e+00 | 4CADM1, OSBPL3, CACNA2D3, ALCAM |
177 |
| GSE18281_SUBCAPSULAR_CORTICAL_REGION_VS_WHOLE_CORTEX_THYMUS_DN | 7.45e-03 | 5.60 | 1.45 | 1.00e+00 | 1.00e+00 | 4CADM1, DPYD, ALCAM, SEMA3E |
198 |
| GSE7460_WT_VS_FOXP3_HET_ACT_TCONV_UP | 7.58e-03 | 5.57 | 1.44 | 1.00e+00 | 1.00e+00 | 4RYR2, CTNND2, ZBTB20, GRID2 |
199 |
| GSE19825_NAIVE_VS_DAY3_EFF_CD8_TCELL_UP | 7.71e-03 | 5.55 | 1.44 | 1.00e+00 | 1.00e+00 | 4NXPH1, ELAVL2, KCND2, FSTL5 |
200 |
| GSE360_L_MAJOR_VS_B_MALAYI_HIGH_DOSE_MAC_UP | 7.71e-03 | 5.55 | 1.44 | 1.00e+00 | 1.00e+00 | 4DPP6, ALCAM, SYN3, FAM155A |
200 |
| GSE41978_ID2_KO_VS_ID2_KO_AND_BIM_KO_KLRG1_LOW_EFFECTOR_CD8_TCELL_UP | 7.71e-03 | 5.55 | 1.44 | 1.00e+00 | 1.00e+00 | 4NXPH1, SOX6, CACNA2D3, ALCAM |
200 |
| GSE41176_WT_VS_TAK1_KO_ANTI_IGM_STIM_BCELL_1H_DN | 7.71e-03 | 5.55 | 1.44 | 1.00e+00 | 1.00e+00 | 4DPYD, DGKB, CNTNAP2, KCND2 |
200 |
| GSE37605_TREG_VS_TCONV_C57BL6_FOXP3_FUSION_GFP_UP | 1.61e-02 | 6.04 | 1.19 | 1.00e+00 | 1.00e+00 | 3RGS7, CACNA2D3, FAM155A |
135 |
| GSE13522_WT_VS_IFNG_KO_SKIN_UP | 1.70e-02 | 5.91 | 1.16 | 1.00e+00 | 1.00e+00 | 3ROBO1, ANKFN1, FSTL5 |
138 |
| GSE12484_HEALTHY_VS_PERIDONTITIS_NEUTROPHILS_UP | 1.73e-02 | 5.87 | 1.15 | 1.00e+00 | 1.00e+00 | 3NXPH1, CADM1, SOX6 |
139 |
| GSE12507_PDC_CELL_LINE_VS_IMMATUE_T_CELL_LINE_DN | 1.90e-02 | 5.66 | 1.11 | 1.00e+00 | 1.00e+00 | 3RGS7, ITGA8, IL7 |
144 |
| GSE12003_4D_VS_8D_CULTURE_MIR223_KO_BM_PROGENITOR_UP | 2.19e-02 | 5.35 | 1.05 | 1.00e+00 | 1.00e+00 | 3CADM1, BRINP3, KCND2 |
152 |
| GSE40274_FOXP3_VS_FOXP3_AND_XBP1_TRANSDUCED_ACTIVATED_CD4_TCELL_DN | 2.45e-02 | 5.11 | 1.01 | 1.00e+00 | 1.00e+00 | 3ALCAM, RAPGEF5, FSTL5 |
159 |
| GSE4590_PRE_BCELL_VS_LARGE_PRE_BCELL_UP | 2.49e-02 | 5.08 | 1.00 | 1.00e+00 | 1.00e+00 | 3DPYD, IL7, GRID2 |
160 |
| GSE6269_E_COLI_VS_STREP_PNEUMO_INF_PBMC_UP | 2.82e-02 | 4.83 | 0.95 | 1.00e+00 | 1.00e+00 | 3PTPRD, ROBO1, AUTS2 |
168 |
| GSE46606_UNSTIM_VS_CD40L_IL2_IL5_3DAY_STIMULATED_IRF4_KO_BCELL_UP | 3.54e-02 | 4.41 | 0.87 | 1.00e+00 | 1.00e+00 | 3ROBO2, CTNND2, CNTNAP2 |
184 |
| GSE17974_IL4_AND_ANTI_IL12_VS_UNTREATED_24H_ACT_CD4_TCELL_UP | 3.64e-02 | 4.36 | 0.86 | 1.00e+00 | 1.00e+00 | 3PTPRD, SORBS2, DLC1 |
186 |
| GSE1791_CTRL_VS_NEUROMEDINU_IN_T_CELL_LINE_3H_UP | 3.74e-02 | 4.31 | 0.85 | 1.00e+00 | 1.00e+00 | 3RGS7, LRRC4C, FSTL5 |
188 |
| GSE45365_BCELL_VS_CD8_TCELL_DN | 3.74e-02 | 4.31 | 0.85 | 1.00e+00 | 1.00e+00 | 3LRRTM4, SOX6, SYN3 |
188 |
Top Ranked Transcription Factors for this Gene Expression Program:
| Gene Symbol | TF Rank | DNA Binding Domain | Motif Status | IUPAC PWM | GTEx | DepMap | Decartes |
|---|---|---|---|---|---|---|---|
| NRG1 | 1 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Cell surface receptor - does not have predicted DBDs |
| EBF1 | 3 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| RGS7 | 10 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| SORBS2 | 21 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| SOX6 | 22 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| ZBTB20 | 34 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| RAPGEF5 | 39 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| KLF12 | 59 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| ZFPM2 | 81 | Yes | Likely to be sequence specific TF | Monomer or homomultimer | No motif | None | FOG-proteins use some of their zinc-fingers to interact with the GATA proteins. Other C2H2 ZFs are probably also capable of binding DNA (PMID: 10329627). |
| TSHZ2 | 89 | Yes | Likely to be sequence specific TF | Monomer or homomultimer | No motif | None | Also contains an atypical homodomain (Uniprot) not identified by Pfam. |
| NTRK1 | 93 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Membrane bound receptor protein |
| ZFHX3 | 94 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| CAMTA1 | 102 | Yes | Likely to be sequence specific TF | Monomer or homomultimer | No motif | None | Binds CGCATTGCG based on EMSA performed in (PMID: 25049392) |
| SOX4 | 113 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | Also binds ssDNA loops. |
| MYRFL | 116 | Yes | Likely to be sequence specific TF | Monomer or homomultimer | No motif | None | Related to other MYRF TFs, which have related PBM motifs |
| ERC1 | 129 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| MACF1 | 141 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Included only because Vasquerizas 2009 includes it with an x. No DBD and no other data to support being a TF. |
| TLE4 | 160 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | TLE4 is a Groucho protein (co-repressor) |
| HIVEP3 | 163 | Yes | Inferred motif | Monomer or homomultimer | In vivo/Misc source | Has a putative AT-hook | Binds to NFKB-like consensus sequence to repress transcription (PMID: 21189157). PWMs for HIVEP1 and 2 in Transfac and Hocomoco are also NFKB-like. |
| RORA | 172 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
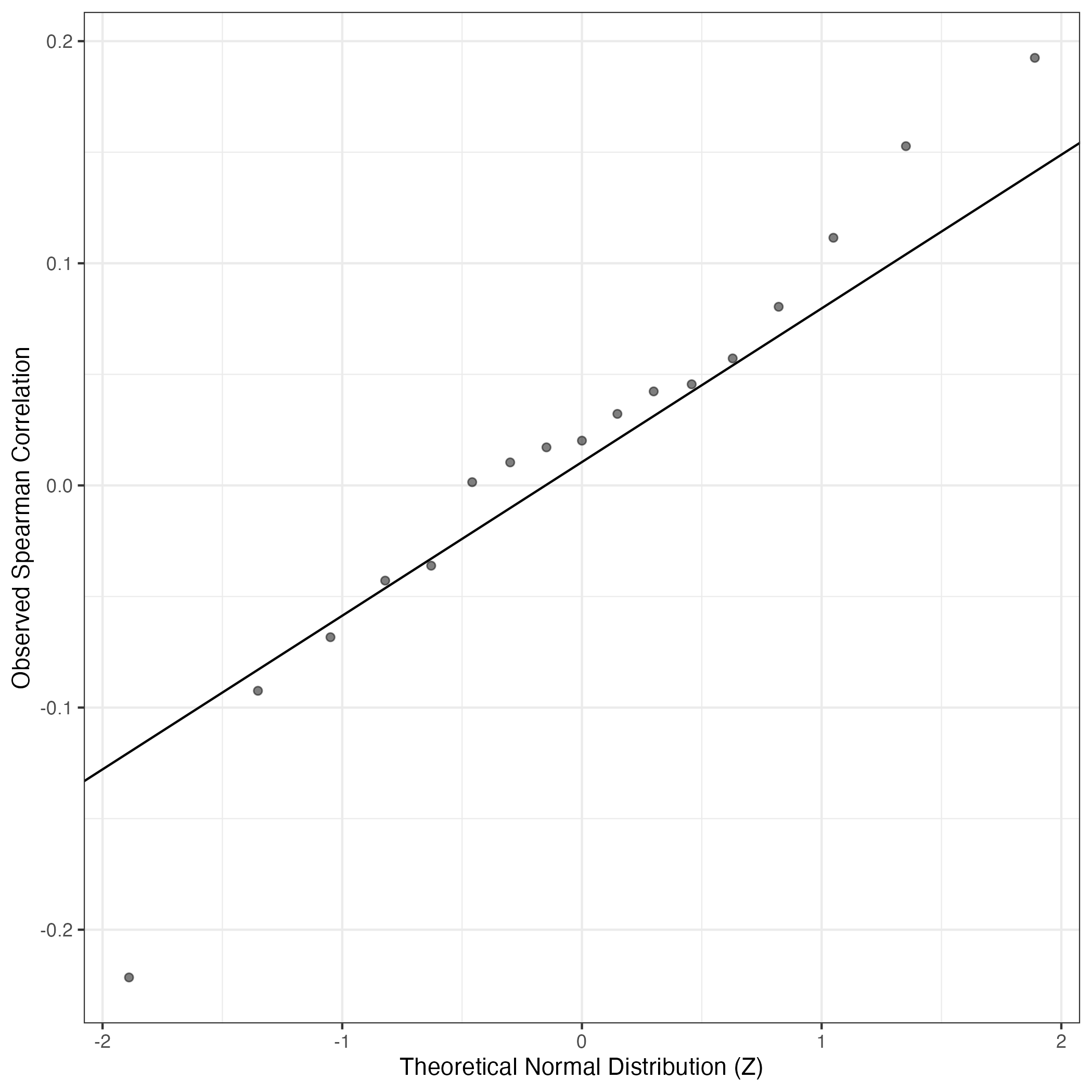
QQ Plot showing correlations with other Gene Expression Programs in this dataset, calculated by Spearman correlation:
Interactive QQ-plot of gene loadings:
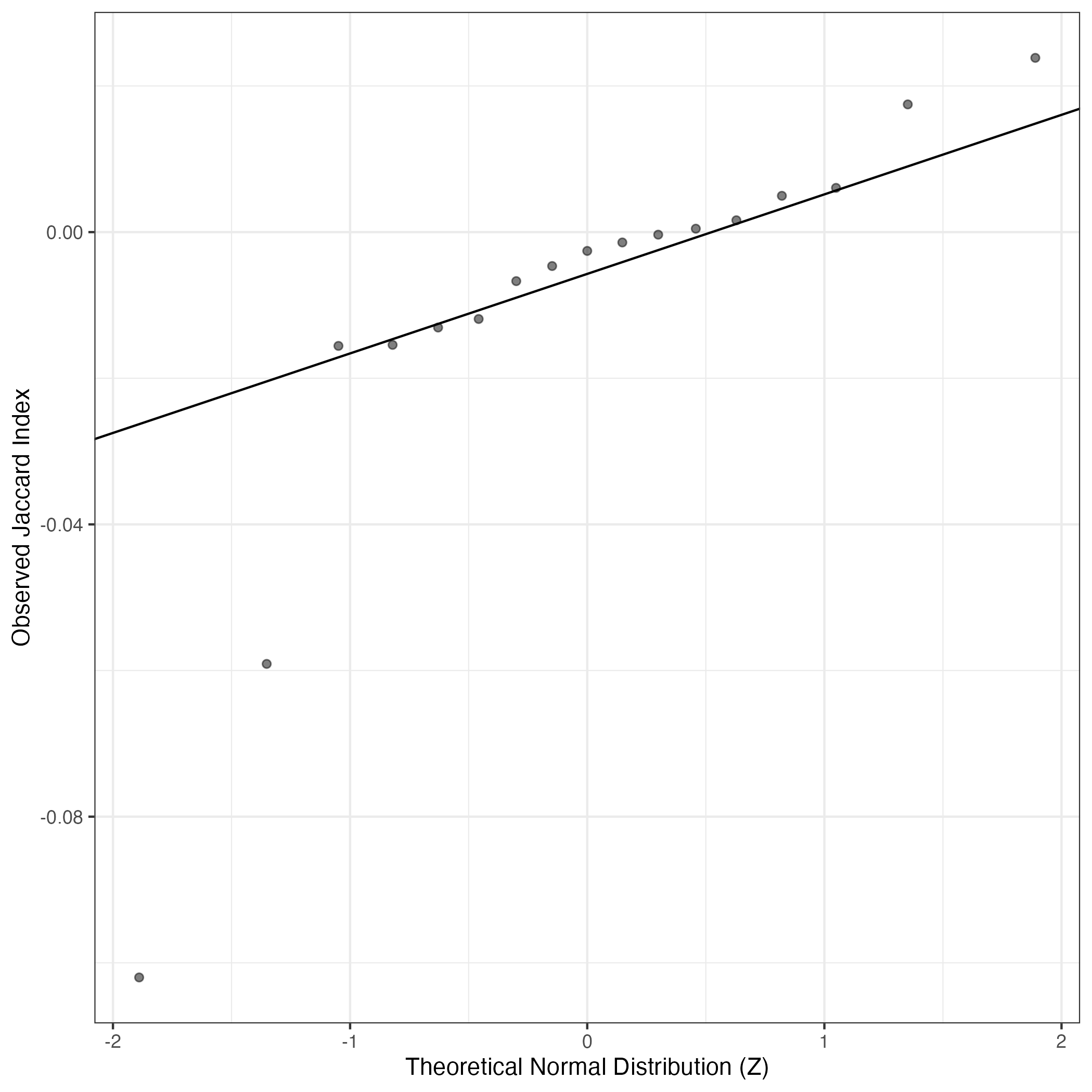
A similar QQ-plot as above, but only for instances where the H value is e.g. > 25, i.e. we are confident that the expression program is active above noise. Agreemenet between these binary vectors is tested using the Jaccard Index, with the P-values calculated by an exact test:
Interactive QQ-plot:
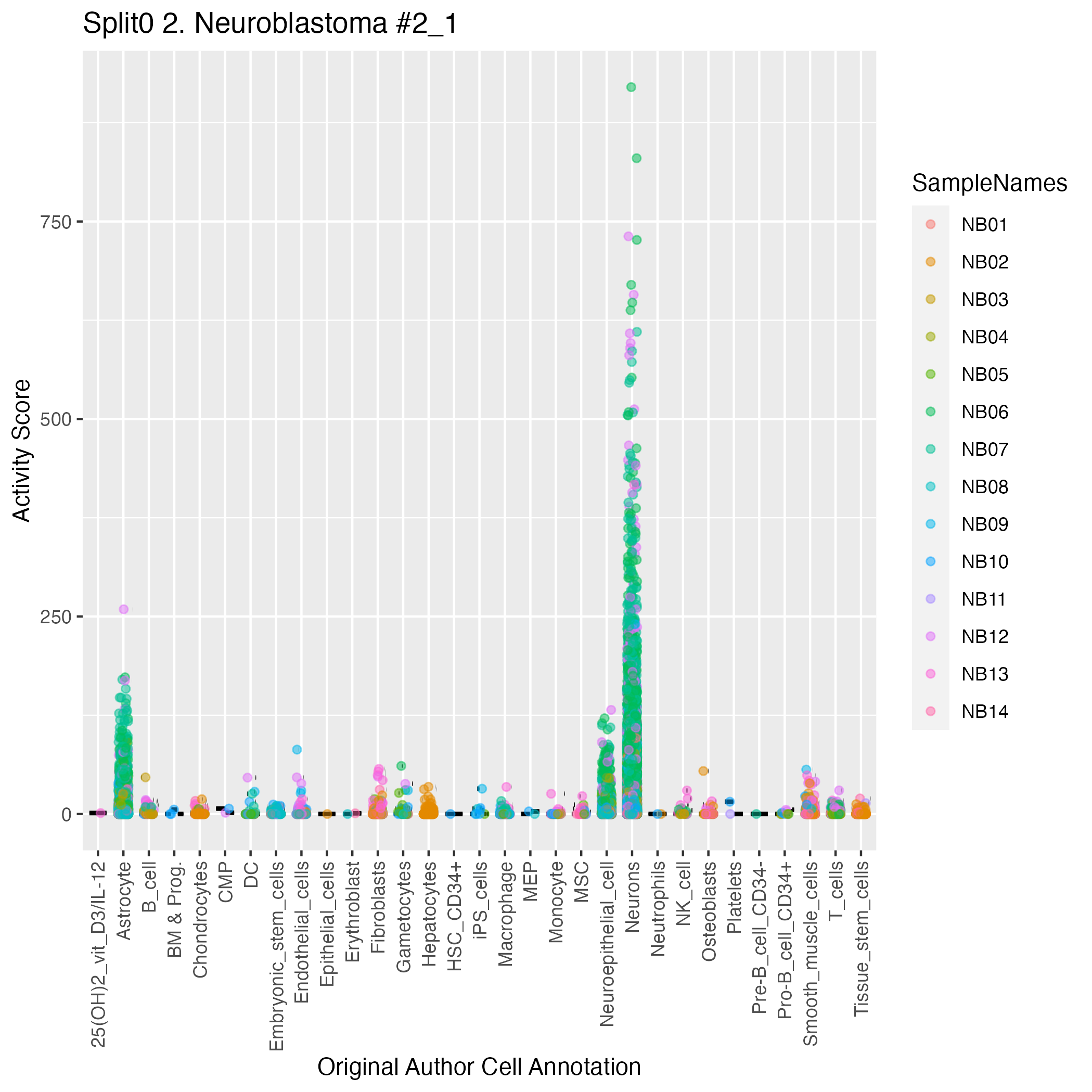
Singler cell type annotations for the top 50 cells on this program.
| Cell ID | Singler label | Singler Delta | Activity Score | Top Singler Raw Scores |
|---|---|---|---|---|
| NB06_CATCGAAAGTTAAGTG-1 | Neurons:adrenal_medulla_cell_line | 0.22 | 920.03 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.34, Neuroepithelial_cell:ESC-derived: 0.33, Neurons:ES_cell-derived_neural_precursor: 0.31, iPS_cells:PDB_1lox-21Puro-20: 0.29, iPS_cells:PDB_1lox-17Puro-10: 0.29, iPS_cells:PDB_1lox-21Puro-26: 0.29, iPS_cells:PDB_1lox-17Puro-5: 0.28, Embryonic_stem_cells: 0.28, iPS_cells:PDB_2lox-22: 0.27 |
| NB06_CACCAGGCACAACGTT-1 | Neurons:adrenal_medulla_cell_line | 0.26 | 830.08 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.46, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.38, Neurons:ES_cell-derived_neural_precursor: 0.34, iPS_cells:PDB_1lox-21Puro-20: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.33, iPS_cells:PDB_1lox-21Puro-26: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.33, iPS_cells:PDB_2lox-22: 0.32, Embryonic_stem_cells: 0.32 |
| NB12_CCGTACTGTCTTCTCG-1 | Neurons:adrenal_medulla_cell_line | 0.25 | 731.16 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.39, Neuroepithelial_cell:ESC-derived: 0.33, Astrocyte:Embryonic_stem_cell-derived: 0.33, Neurons:ES_cell-derived_neural_precursor: 0.3, iPS_cells:PDB_1lox-21Puro-20: 0.29, iPS_cells:PDB_1lox-21Puro-26: 0.29, iPS_cells:PDB_1lox-17Puro-10: 0.29, iPS_cells:PDB_1lox-17Puro-5: 0.28, Embryonic_stem_cells: 0.28, iPS_cells:PDB_2lox-22: 0.27 |
| NB06_CACTCCACAATCTACG-1 | Neurons:adrenal_medulla_cell_line | 0.19 | 726.65 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Neuroepithelial_cell:ESC-derived: 0.31, Astrocyte:Embryonic_stem_cell-derived: 0.31, Neurons:ES_cell-derived_neural_precursor: 0.3, Embryonic_stem_cells: 0.26, iPS_cells:PDB_1lox-17Puro-10: 0.25, iPS_cells:PDB_1lox-21Puro-20: 0.25, iPS_cells:PDB_1lox-21Puro-26: 0.25, MSC: 0.25, Tissue_stem_cells:CD326-CD56+: 0.25 |
| NB06_TCACAAGCACAGCGTC-1 | Neurons:adrenal_medulla_cell_line | 0.25 | 669.83 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.4, Astrocyte:Embryonic_stem_cell-derived: 0.33, Neuroepithelial_cell:ESC-derived: 0.33, Neurons:ES_cell-derived_neural_precursor: 0.3, iPS_cells:PDB_1lox-21Puro-20: 0.28, iPS_cells:PDB_1lox-17Puro-10: 0.28, iPS_cells:PDB_1lox-21Puro-26: 0.28, iPS_cells:PDB_1lox-17Puro-5: 0.28, Embryonic_stem_cells: 0.27, iPS_cells:PDB_2lox-22: 0.27 |
| NB12_ATAGACCTCAGCTGGC-1 | Neurons:adrenal_medulla_cell_line | 0.24 | 657.26 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.4, Neuroepithelial_cell:ESC-derived: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.34, Neurons:ES_cell-derived_neural_precursor: 0.33, iPS_cells:PDB_1lox-21Puro-20: 0.31, iPS_cells:PDB_1lox-21Puro-26: 0.31, iPS_cells:PDB_1lox-17Puro-10: 0.31, iPS_cells:PDB_1lox-17Puro-5: 0.31, Embryonic_stem_cells: 0.3, iPS_cells:PDB_2lox-22: 0.3 |
| NB06_CACCACTAGATATACG-1 | Neurons:adrenal_medulla_cell_line | 0.26 | 647.35 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.5, Neuroepithelial_cell:ESC-derived: 0.43, Astrocyte:Embryonic_stem_cell-derived: 0.42, Neurons:ES_cell-derived_neural_precursor: 0.39, iPS_cells:PDB_1lox-21Puro-20: 0.38, iPS_cells:PDB_1lox-17Puro-10: 0.38, iPS_cells:PDB_1lox-21Puro-26: 0.38, iPS_cells:PDB_1lox-17Puro-5: 0.38, iPS_cells:PDB_2lox-22: 0.37, Embryonic_stem_cells: 0.37 |
| NB06_CGTTCTGAGGCTCATT-1 | Neurons:adrenal_medulla_cell_line | 0.22 | 637.64 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.32, Astrocyte:Embryonic_stem_cell-derived: 0.26, Neuroepithelial_cell:ESC-derived: 0.25, Neurons:ES_cell-derived_neural_precursor: 0.24, iPS_cells:PDB_1lox-17Puro-10: 0.21, iPS_cells:PDB_1lox-21Puro-20: 0.21, iPS_cells:PDB_1lox-21Puro-26: 0.21, Embryonic_stem_cells: 0.2, iPS_cells:PDB_1lox-17Puro-5: 0.2, iPS_cells:skin_fibroblast-derived: 0.2 |
| NB07_CGAACATAGTGCCATT-1 | Neurons:adrenal_medulla_cell_line | 0.23 | 610.34 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.36, Astrocyte:Embryonic_stem_cell-derived: 0.3, Neuroepithelial_cell:ESC-derived: 0.3, Neurons:ES_cell-derived_neural_precursor: 0.28, iPS_cells:PDB_1lox-17Puro-10: 0.26, iPS_cells:PDB_1lox-21Puro-20: 0.26, iPS_cells:PDB_1lox-21Puro-26: 0.26, iPS_cells:PDB_1lox-17Puro-5: 0.26, Embryonic_stem_cells: 0.25, iPS_cells:PDB_2lox-22: 0.24 |
| NB12_CTACGTCTCTCTGCTG-1 | Neurons:adrenal_medulla_cell_line | 0.19 | 608.34 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.31, Neuroepithelial_cell:ESC-derived: 0.27, Astrocyte:Embryonic_stem_cell-derived: 0.27, Neurons:ES_cell-derived_neural_precursor: 0.27, iPS_cells:PDB_1lox-21Puro-20: 0.24, iPS_cells:PDB_1lox-17Puro-10: 0.24, iPS_cells:PDB_1lox-17Puro-5: 0.24, iPS_cells:PDB_1lox-21Puro-26: 0.24, iPS_cells:CRL2097_foreskin: 0.23, Embryonic_stem_cells: 0.23 |
| NB12_TGAGGGAGTAAATGAC-1 | Neurons:adrenal_medulla_cell_line | 0.23 | 596.19 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.35, Neuroepithelial_cell:ESC-derived: 0.29, Astrocyte:Embryonic_stem_cell-derived: 0.28, Neurons:ES_cell-derived_neural_precursor: 0.27, iPS_cells:PDB_1lox-21Puro-20: 0.26, iPS_cells:PDB_1lox-17Puro-10: 0.25, iPS_cells:PDB_1lox-21Puro-26: 0.25, iPS_cells:PDB_1lox-17Puro-5: 0.25, Embryonic_stem_cells: 0.24, iPS_cells:PDB_2lox-22: 0.24 |
| NB12_TTTGCGCTCAGGATCT-1 | Neurons:adrenal_medulla_cell_line | 0.22 | 589.68 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Neuroepithelial_cell:ESC-derived: 0.29, Astrocyte:Embryonic_stem_cell-derived: 0.29, Neurons:ES_cell-derived_neural_precursor: 0.28, iPS_cells:PDB_1lox-21Puro-20: 0.25, Embryonic_stem_cells: 0.25, iPS_cells:PDB_1lox-17Puro-10: 0.25, iPS_cells:PDB_1lox-21Puro-26: 0.25, iPS_cells:PDB_1lox-17Puro-5: 0.24, iPS_cells:skin_fibroblast-derived: 0.24 |
| NB07_CTAATGGGTTACGTCA-1 | Neurons:adrenal_medulla_cell_line | 0.23 | 585.99 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Astrocyte:Embryonic_stem_cell-derived: 0.27, Neuroepithelial_cell:ESC-derived: 0.27, Neurons:ES_cell-derived_neural_precursor: 0.26, iPS_cells:PDB_1lox-21Puro-20: 0.23, iPS_cells:PDB_1lox-17Puro-10: 0.23, iPS_cells:PDB_1lox-21Puro-26: 0.23, iPS_cells:PDB_1lox-17Puro-5: 0.23, Embryonic_stem_cells: 0.22, iPS_cells:skin_fibroblast-derived: 0.21 |
| NB12_ACACCAAAGGACCACA-1 | Neurons:adrenal_medulla_cell_line | 0.25 | 580.73 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.4, Neuroepithelial_cell:ESC-derived: 0.34, Astrocyte:Embryonic_stem_cell-derived: 0.33, Neurons:ES_cell-derived_neural_precursor: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.31, iPS_cells:PDB_1lox-21Puro-20: 0.31, iPS_cells:PDB_1lox-21Puro-26: 0.31, iPS_cells:PDB_1lox-17Puro-5: 0.31, iPS_cells:PDB_2lox-22: 0.3, Embryonic_stem_cells: 0.3 |
| NB07_AAGACCTAGACTAAGT-1 | Neurons:adrenal_medulla_cell_line | 0.20 | 572.13 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.32, Neuroepithelial_cell:ESC-derived: 0.27, Astrocyte:Embryonic_stem_cell-derived: 0.27, Neurons:ES_cell-derived_neural_precursor: 0.25, iPS_cells:PDB_1lox-21Puro-20: 0.24, iPS_cells:PDB_1lox-17Puro-10: 0.24, iPS_cells:PDB_1lox-21Puro-26: 0.24, iPS_cells:PDB_1lox-17Puro-5: 0.23, Embryonic_stem_cells: 0.23, iPS_cells:PDB_2lox-22: 0.23 |
| NB06_TCTCTAACACTGAAGG-1 | Neurons:adrenal_medulla_cell_line | 0.22 | 552.39 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.36, Astrocyte:Embryonic_stem_cell-derived: 0.29, Neuroepithelial_cell:ESC-derived: 0.28, Neurons:ES_cell-derived_neural_precursor: 0.25, iPS_cells:PDB_1lox-21Puro-20: 0.24, iPS_cells:PDB_1lox-17Puro-10: 0.24, iPS_cells:PDB_1lox-21Puro-26: 0.23, Embryonic_stem_cells: 0.23, iPS_cells:PDB_1lox-17Puro-5: 0.23, iPS_cells:PDB_2lox-22: 0.22 |
| NB07_GGAACTTCAATAAGCA-1 | Neurons:adrenal_medulla_cell_line | 0.22 | 549.25 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.37, Astrocyte:Embryonic_stem_cell-derived: 0.31, Neuroepithelial_cell:ESC-derived: 0.3, Neurons:ES_cell-derived_neural_precursor: 0.28, iPS_cells:PDB_1lox-21Puro-20: 0.26, iPS_cells:PDB_1lox-21Puro-26: 0.26, iPS_cells:PDB_1lox-17Puro-10: 0.26, iPS_cells:PDB_1lox-17Puro-5: 0.26, Embryonic_stem_cells: 0.25, iPS_cells:PDB_2lox-22: 0.25 |
| NB07_CCACCTACACCCATGG-1 | Neurons:adrenal_medulla_cell_line | 0.24 | 545.89 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.38, Neuroepithelial_cell:ESC-derived: 0.32, Astrocyte:Embryonic_stem_cell-derived: 0.3, Neurons:ES_cell-derived_neural_precursor: 0.3, iPS_cells:PDB_1lox-21Puro-20: 0.29, iPS_cells:PDB_1lox-17Puro-10: 0.28, iPS_cells:PDB_1lox-21Puro-26: 0.28, iPS_cells:PDB_1lox-17Puro-5: 0.28, iPS_cells:PDB_2lox-22: 0.27, iPS_cells:PDB_2lox-21: 0.27 |
| NB12_CCTACCAGTTTGACTG-1 | Neurons:adrenal_medulla_cell_line | 0.20 | 512.27 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Astrocyte:Embryonic_stem_cell-derived: 0.27, Neuroepithelial_cell:ESC-derived: 0.27, Neurons:ES_cell-derived_neural_precursor: 0.25, iPS_cells:PDB_1lox-17Puro-10: 0.22, iPS_cells:PDB_1lox-21Puro-20: 0.22, iPS_cells:PDB_1lox-21Puro-26: 0.22, iPS_cells:PDB_1lox-17Puro-5: 0.22, Embryonic_stem_cells: 0.22, iPS_cells:skin_fibroblast-derived: 0.21 |
| NB06_ATCTACTAGTTGAGTA-1 | Neurons:adrenal_medulla_cell_line | 0.21 | 508.73 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.32, Neuroepithelial_cell:ESC-derived: 0.28, Astrocyte:Embryonic_stem_cell-derived: 0.27, Neurons:ES_cell-derived_neural_precursor: 0.26, iPS_cells:PDB_1lox-21Puro-20: 0.24, iPS_cells:PDB_1lox-21Puro-26: 0.23, iPS_cells:PDB_1lox-17Puro-10: 0.23, Embryonic_stem_cells: 0.23, iPS_cells:PDB_1lox-17Puro-5: 0.23, iPS_cells:PDB_2lox-22: 0.22 |
| NB07_TCAATCTTCGCACTCT-1 | Neurons:adrenal_medulla_cell_line | 0.19 | 508.25 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.26, Astrocyte:Embryonic_stem_cell-derived: 0.21, Neuroepithelial_cell:ESC-derived: 0.2, Neurons:ES_cell-derived_neural_precursor: 0.2, iPS_cells:PDB_1lox-21Puro-20: 0.17, iPS_cells:PDB_1lox-17Puro-10: 0.17, iPS_cells:PDB_1lox-21Puro-26: 0.17, iPS_cells:PDB_1lox-17Puro-5: 0.16, Embryonic_stem_cells: 0.16, iPS_cells:PDB_2lox-22: 0.15 |
| NB07_AGCTTGATCTGTACGA-1 | Neurons:adrenal_medulla_cell_line | 0.23 | 505.06 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Neuroepithelial_cell:ESC-derived: 0.27, Astrocyte:Embryonic_stem_cell-derived: 0.27, Neurons:ES_cell-derived_neural_precursor: 0.24, iPS_cells:PDB_1lox-21Puro-20: 0.23, iPS_cells:PDB_1lox-17Puro-10: 0.23, iPS_cells:PDB_1lox-21Puro-26: 0.22, iPS_cells:PDB_1lox-17Puro-5: 0.22, Embryonic_stem_cells: 0.22, iPS_cells:PDB_2lox-22: 0.21 |
| NB06_CAGTAACGTAAACACA-1 | Neurons:adrenal_medulla_cell_line | 0.24 | 504.43 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.46, Neuroepithelial_cell:ESC-derived: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.39, Neurons:ES_cell-derived_neural_precursor: 0.36, iPS_cells:PDB_1lox-21Puro-20: 0.33, Embryonic_stem_cells: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.33, iPS_cells:PDB_1lox-21Puro-26: 0.33, Tissue_stem_cells:CD326-CD56+: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.33 |
| NB12_CCTATTAGTACTTCTT-1 | Neurons:adrenal_medulla_cell_line | 0.21 | 466.60 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.31, Neuroepithelial_cell:ESC-derived: 0.26, Astrocyte:Embryonic_stem_cell-derived: 0.26, Neurons:ES_cell-derived_neural_precursor: 0.24, iPS_cells:PDB_1lox-21Puro-20: 0.22, iPS_cells:PDB_1lox-21Puro-26: 0.22, iPS_cells:PDB_1lox-17Puro-10: 0.22, iPS_cells:PDB_1lox-17Puro-5: 0.22, Embryonic_stem_cells: 0.22, iPS_cells:skin_fibroblast-derived: 0.21 |
| NB06_CTCATTATCTGACCTC-1 | Neurons:adrenal_medulla_cell_line | 0.19 | 462.83 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.29, Astrocyte:Embryonic_stem_cell-derived: 0.23, Neuroepithelial_cell:ESC-derived: 0.22, Neurons:ES_cell-derived_neural_precursor: 0.21, iPS_cells:PDB_1lox-21Puro-20: 0.19, iPS_cells:PDB_1lox-17Puro-10: 0.19, iPS_cells:PDB_1lox-21Puro-26: 0.19, iPS_cells:PDB_1lox-17Puro-5: 0.19, Embryonic_stem_cells: 0.18, iPS_cells:PDB_2lox-22: 0.18 |
| NB07_TTATGCTAGTTACGGG-1 | Neurons:adrenal_medulla_cell_line | 0.20 | 456.30 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Neuroepithelial_cell:ESC-derived: 0.26, Astrocyte:Embryonic_stem_cell-derived: 0.25, Neurons:ES_cell-derived_neural_precursor: 0.23, iPS_cells:PDB_1lox-17Puro-10: 0.21, iPS_cells:PDB_1lox-21Puro-20: 0.21, iPS_cells:PDB_1lox-17Puro-5: 0.21, iPS_cells:PDB_1lox-21Puro-26: 0.21, Embryonic_stem_cells: 0.2, iPS_cells:PDB_2lox-22: 0.2 |
| NB07_GATCAGTTCTAAGCCA-1 | Neurons:adrenal_medulla_cell_line | 0.23 | 453.46 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Astrocyte:Embryonic_stem_cell-derived: 0.26, Neuroepithelial_cell:ESC-derived: 0.26, Neurons:ES_cell-derived_neural_precursor: 0.25, iPS_cells:PDB_1lox-21Puro-20: 0.23, iPS_cells:PDB_1lox-21Puro-26: 0.23, iPS_cells:PDB_1lox-17Puro-10: 0.22, iPS_cells:PDB_1lox-17Puro-5: 0.22, Embryonic_stem_cells: 0.22, iPS_cells:PDB_2lox-22: 0.22 |
| NB12_GAACATCTCTTCAACT-1 | Neurons:adrenal_medulla_cell_line | 0.22 | 448.09 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Neuroepithelial_cell:ESC-derived: 0.29, Astrocyte:Embryonic_stem_cell-derived: 0.28, Neurons:ES_cell-derived_neural_precursor: 0.27, iPS_cells:PDB_1lox-21Puro-20: 0.25, iPS_cells:PDB_1lox-21Puro-26: 0.25, iPS_cells:PDB_1lox-17Puro-10: 0.25, iPS_cells:PDB_1lox-17Puro-5: 0.24, Embryonic_stem_cells: 0.24, iPS_cells:skin_fibroblast-derived: 0.23 |
| NB07_TTGCGTCTCCTTTCTC-1 | Neurons:adrenal_medulla_cell_line | 0.21 | 445.65 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Astrocyte:Embryonic_stem_cell-derived: 0.28, Neuroepithelial_cell:ESC-derived: 0.27, Neurons:ES_cell-derived_neural_precursor: 0.26, iPS_cells:PDB_1lox-21Puro-20: 0.24, iPS_cells:PDB_1lox-21Puro-26: 0.24, iPS_cells:PDB_1lox-17Puro-10: 0.23, iPS_cells:PDB_1lox-17Puro-5: 0.23, iPS_cells:PDB_2lox-22: 0.22, Embryonic_stem_cells: 0.22 |
| NB06_AGGGATGAGTCAAGCG-1 | Neurons:adrenal_medulla_cell_line | 0.21 | 444.03 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.29, Astrocyte:Embryonic_stem_cell-derived: 0.23, Neuroepithelial_cell:ESC-derived: 0.22, Neurons:ES_cell-derived_neural_precursor: 0.19, iPS_cells:PDB_1lox-21Puro-20: 0.17, iPS_cells:PDB_1lox-21Puro-26: 0.17, iPS_cells:PDB_1lox-17Puro-10: 0.17, Embryonic_stem_cells: 0.17, iPS_cells:PDB_1lox-17Puro-5: 0.17, iPS_cells:PDB_2lox-22: 0.16 |
| NB07_TTCTCCTAGTGACATA-1 | Neurons:adrenal_medulla_cell_line | 0.23 | 441.50 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.34, Neuroepithelial_cell:ESC-derived: 0.29, Astrocyte:Embryonic_stem_cell-derived: 0.27, Neurons:ES_cell-derived_neural_precursor: 0.26, iPS_cells:PDB_1lox-21Puro-20: 0.25, iPS_cells:PDB_1lox-17Puro-10: 0.25, iPS_cells:PDB_1lox-21Puro-26: 0.25, iPS_cells:PDB_1lox-17Puro-5: 0.24, Embryonic_stem_cells: 0.24, iPS_cells:PDB_2lox-22: 0.23 |
| NB12_GTACTTTTCGCTTGTC-1 | Neurons:adrenal_medulla_cell_line | 0.20 | 440.89 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.31, Neuroepithelial_cell:ESC-derived: 0.26, Astrocyte:Embryonic_stem_cell-derived: 0.25, Neurons:ES_cell-derived_neural_precursor: 0.24, iPS_cells:PDB_1lox-21Puro-20: 0.22, iPS_cells:PDB_1lox-21Puro-26: 0.22, iPS_cells:PDB_1lox-17Puro-10: 0.22, iPS_cells:PDB_1lox-17Puro-5: 0.22, Embryonic_stem_cells: 0.22, iPS_cells:PDB_2lox-22: 0.21 |
| NB07_AAATGCCTCGCACTCT-1 | Neurons:adrenal_medulla_cell_line | 0.19 | 436.81 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Astrocyte:Embryonic_stem_cell-derived: 0.25, Neuroepithelial_cell:ESC-derived: 0.24, Neurons:ES_cell-derived_neural_precursor: 0.22, iPS_cells:PDB_1lox-21Puro-20: 0.21, iPS_cells:PDB_1lox-17Puro-10: 0.21, iPS_cells:PDB_1lox-21Puro-26: 0.21, iPS_cells:PDB_1lox-17Puro-5: 0.21, iPS_cells:PDB_2lox-22: 0.2, Embryonic_stem_cells: 0.2 |
| NB06_ACCTTTACAAATTGCC-1 | Neurons:adrenal_medulla_cell_line | 0.23 | 432.84 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.39, Neuroepithelial_cell:ESC-derived: 0.32, Astrocyte:Embryonic_stem_cell-derived: 0.32, Neurons:ES_cell-derived_neural_precursor: 0.29, iPS_cells:PDB_1lox-21Puro-20: 0.28, iPS_cells:PDB_1lox-21Puro-26: 0.28, iPS_cells:PDB_1lox-17Puro-10: 0.28, iPS_cells:PDB_1lox-17Puro-5: 0.28, Embryonic_stem_cells: 0.27, iPS_cells:PDB_2lox-22: 0.27 |
| NB07_CACATAGTCCTACAGA-1 | Neurons:adrenal_medulla_cell_line | 0.21 | 427.33 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Neuroepithelial_cell:ESC-derived: 0.24, Astrocyte:Embryonic_stem_cell-derived: 0.24, Neurons:ES_cell-derived_neural_precursor: 0.23, iPS_cells:PDB_1lox-21Puro-20: 0.21, iPS_cells:PDB_1lox-17Puro-10: 0.21, iPS_cells:PDB_1lox-21Puro-26: 0.21, iPS_cells:PDB_1lox-17Puro-5: 0.2, Embryonic_stem_cells: 0.2, iPS_cells:PDB_2lox-22: 0.2 |
| NB06_GATCTAGAGACGCAAC-1 | Neurons:adrenal_medulla_cell_line | 0.23 | 425.44 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.34, Neuroepithelial_cell:ESC-derived: 0.28, Astrocyte:Embryonic_stem_cell-derived: 0.27, Neurons:ES_cell-derived_neural_precursor: 0.25, iPS_cells:PDB_1lox-21Puro-20: 0.23, Embryonic_stem_cells: 0.23, iPS_cells:PDB_1lox-17Puro-10: 0.23, iPS_cells:PDB_1lox-21Puro-26: 0.23, iPS_cells:PDB_1lox-17Puro-5: 0.22, iPS_cells:PDB_2lox-22: 0.22 |
| NB07_CGCTATCCAAGCCGCT-1 | Neurons:adrenal_medulla_cell_line | 0.20 | 419.72 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.29, Neuroepithelial_cell:ESC-derived: 0.23, Astrocyte:Embryonic_stem_cell-derived: 0.23, Neurons:ES_cell-derived_neural_precursor: 0.22, iPS_cells:PDB_1lox-21Puro-20: 0.2, iPS_cells:PDB_1lox-21Puro-26: 0.2, iPS_cells:PDB_1lox-17Puro-5: 0.2, iPS_cells:PDB_1lox-17Puro-10: 0.2, Embryonic_stem_cells: 0.2, iPS_cells:PDB_2lox-22: 0.19 |
| NB12_TTTGCGCGTCCAGTTA-1 | Neurons:adrenal_medulla_cell_line | 0.21 | 417.35 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.32, Astrocyte:Embryonic_stem_cell-derived: 0.26, Neuroepithelial_cell:ESC-derived: 0.26, Neurons:ES_cell-derived_neural_precursor: 0.25, iPS_cells:PDB_1lox-21Puro-20: 0.22, iPS_cells:PDB_1lox-17Puro-10: 0.22, iPS_cells:PDB_1lox-21Puro-26: 0.22, iPS_cells:PDB_1lox-17Puro-5: 0.21, Embryonic_stem_cells: 0.21, iPS_cells:PDB_2lox-22: 0.21 |
| NB13_CTAACTTAGCCGCCTA-1 | Neurons:adrenal_medulla_cell_line | 0.23 | 416.88 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.42, Astrocyte:Embryonic_stem_cell-derived: 0.34, Neuroepithelial_cell:ESC-derived: 0.33, Neurons:ES_cell-derived_neural_precursor: 0.31, iPS_cells:PDB_1lox-21Puro-20: 0.3, iPS_cells:PDB_1lox-21Puro-26: 0.3, iPS_cells:PDB_1lox-17Puro-10: 0.3, iPS_cells:PDB_1lox-17Puro-5: 0.3, Embryonic_stem_cells: 0.29, iPS_cells:PDB_2lox-22: 0.29 |
| NB07_ATTTCTGGTTATCACG-1 | Neurons:adrenal_medulla_cell_line | 0.22 | 413.67 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.29, Neuroepithelial_cell:ESC-derived: 0.24, Astrocyte:Embryonic_stem_cell-derived: 0.23, Neurons:ES_cell-derived_neural_precursor: 0.23, iPS_cells:PDB_1lox-21Puro-20: 0.21, iPS_cells:PDB_1lox-17Puro-10: 0.21, iPS_cells:PDB_1lox-21Puro-26: 0.21, iPS_cells:PDB_1lox-17Puro-5: 0.21, Embryonic_stem_cells: 0.2, iPS_cells:PDB_2lox-22: 0.19 |
| NB12_GGCGTGTGTCTTTCAT-1 | Neurons:adrenal_medulla_cell_line | 0.22 | 406.95 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Neuroepithelial_cell:ESC-derived: 0.26, Astrocyte:Embryonic_stem_cell-derived: 0.25, Neurons:ES_cell-derived_neural_precursor: 0.24, iPS_cells:PDB_1lox-21Puro-20: 0.22, iPS_cells:PDB_1lox-17Puro-10: 0.21, iPS_cells:PDB_1lox-21Puro-26: 0.21, iPS_cells:PDB_1lox-17Puro-5: 0.21, Embryonic_stem_cells: 0.21, iPS_cells:PDB_2lox-22: 0.2 |
| NB07_ATGTGTGTCCGTAGTA-1 | Neurons:adrenal_medulla_cell_line | 0.19 | 404.21 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.27, Neuroepithelial_cell:ESC-derived: 0.22, Astrocyte:Embryonic_stem_cell-derived: 0.22, Neurons:ES_cell-derived_neural_precursor: 0.21, iPS_cells:PDB_1lox-21Puro-20: 0.19, iPS_cells:PDB_1lox-17Puro-10: 0.19, iPS_cells:PDB_1lox-21Puro-26: 0.19, iPS_cells:PDB_1lox-17Puro-5: 0.19, Embryonic_stem_cells: 0.18, iPS_cells:PDB_2lox-22: 0.18 |
| NB07_CGCGGTAGTCCGTTAA-1 | Neurons:adrenal_medulla_cell_line | 0.25 | 394.26 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.42, Neuroepithelial_cell:ESC-derived: 0.36, Astrocyte:Embryonic_stem_cell-derived: 0.34, Neurons:ES_cell-derived_neural_precursor: 0.33, iPS_cells:PDB_1lox-21Puro-20: 0.32, iPS_cells:PDB_1lox-17Puro-10: 0.32, iPS_cells:PDB_1lox-21Puro-26: 0.32, iPS_cells:PDB_1lox-17Puro-5: 0.31, iPS_cells:PDB_2lox-22: 0.31, Embryonic_stem_cells: 0.3 |
| NB12_TGGACGCCAATTGCTG-1 | Neurons:adrenal_medulla_cell_line | 0.21 | 389.24 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.31, Astrocyte:Embryonic_stem_cell-derived: 0.26, Neuroepithelial_cell:ESC-derived: 0.26, Neurons:ES_cell-derived_neural_precursor: 0.24, iPS_cells:PDB_1lox-21Puro-20: 0.22, iPS_cells:PDB_1lox-17Puro-10: 0.22, iPS_cells:PDB_1lox-21Puro-26: 0.21, iPS_cells:PDB_1lox-17Puro-5: 0.21, Embryonic_stem_cells: 0.21, iPS_cells:PDB_2lox-22: 0.21 |
| NB06_CAGATCAAGAAGCCCA-1 | Neurons:adrenal_medulla_cell_line | 0.22 | 387.15 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Astrocyte:Embryonic_stem_cell-derived: 0.28, Neuroepithelial_cell:ESC-derived: 0.27, Neurons:ES_cell-derived_neural_precursor: 0.25, iPS_cells:PDB_1lox-21Puro-20: 0.23, iPS_cells:PDB_1lox-17Puro-10: 0.23, iPS_cells:PDB_1lox-21Puro-26: 0.23, iPS_cells:PDB_1lox-17Puro-5: 0.22, Embryonic_stem_cells: 0.22, iPS_cells:PDB_2lox-22: 0.21 |
| NB06_CCCAATCCATGCAATC-1 | Neurons:adrenal_medulla_cell_line | 0.20 | 381.47 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.27, Neuroepithelial_cell:ESC-derived: 0.21, Astrocyte:Embryonic_stem_cell-derived: 0.21, Neurons:ES_cell-derived_neural_precursor: 0.2, iPS_cells:PDB_1lox-21Puro-20: 0.18, iPS_cells:PDB_1lox-17Puro-10: 0.18, iPS_cells:PDB_1lox-21Puro-26: 0.18, iPS_cells:PDB_1lox-17Puro-5: 0.18, iPS_cells:skin_fibroblast-derived: 0.17, Embryonic_stem_cells: 0.17 |
| NB06_ACTTTCAAGGGTTTCT-1 | Neurons:adrenal_medulla_cell_line | 0.21 | 379.98 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.29, Neuroepithelial_cell:ESC-derived: 0.24, Astrocyte:Embryonic_stem_cell-derived: 0.24, Neurons:ES_cell-derived_neural_precursor: 0.22, iPS_cells:PDB_1lox-21Puro-20: 0.2, Embryonic_stem_cells: 0.2, iPS_cells:PDB_1lox-17Puro-10: 0.2, iPS_cells:PDB_1lox-21Puro-26: 0.19, iPS_cells:PDB_1lox-17Puro-5: 0.19, iPS_cells:PDB_2lox-22: 0.19 |
| NB06_ACTATCTGTACAGCAG-1 | Neurons:adrenal_medulla_cell_line | 0.22 | 377.31 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.34, Astrocyte:Embryonic_stem_cell-derived: 0.28, Neuroepithelial_cell:ESC-derived: 0.27, Neurons:ES_cell-derived_neural_precursor: 0.25, iPS_cells:PDB_1lox-21Puro-20: 0.23, iPS_cells:PDB_1lox-17Puro-10: 0.23, iPS_cells:PDB_1lox-21Puro-26: 0.23, Embryonic_stem_cells: 0.22, iPS_cells:PDB_1lox-17Puro-5: 0.22, iPS_cells:PDB_2lox-22: 0.22 |
| NB07_CCTCAGTTCGGTGTCG-1 | Neurons:adrenal_medulla_cell_line | 0.18 | 373.79 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.28, Astrocyte:Embryonic_stem_cell-derived: 0.24, Neuroepithelial_cell:ESC-derived: 0.24, Neurons:ES_cell-derived_neural_precursor: 0.24, iPS_cells:PDB_1lox-17Puro-10: 0.21, iPS_cells:PDB_1lox-21Puro-20: 0.21, iPS_cells:PDB_1lox-17Puro-5: 0.2, iPS_cells:PDB_1lox-21Puro-26: 0.2, Embryonic_stem_cells: 0.2, iPS_cells:PDB_2lox-22: 0.19 |
| NB12_TTATGCTAGCTGTCTA-1 | Neurons:adrenal_medulla_cell_line | 0.20 | 373.14 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Neuroepithelial_cell:ESC-derived: 0.26, Astrocyte:Embryonic_stem_cell-derived: 0.25, Neurons:ES_cell-derived_neural_precursor: 0.24, iPS_cells:PDB_1lox-21Puro-20: 0.23, iPS_cells:PDB_1lox-21Puro-26: 0.22, iPS_cells:PDB_1lox-17Puro-10: 0.22, Embryonic_stem_cells: 0.22, iPS_cells:PDB_1lox-17Puro-5: 0.22, iPS_cells:skin_fibroblast-derived: 0.22 |
Below shows the significant enrichments of this GEP for literature curated gene lists
This data was procured from existing single cell RNA-seq maps of neuroblastoma or related relevant data.
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by the P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Adrenergic mature (olsen)
Stated on pages 6 and 7 of the main text of Olsen et al. https://www.biorxiv.org/content/10.1101/2020.05.04.077057v1 - it is argued that upon reanalysis of the adrenergic neuroblastoma cells, there is further heterogeneity, grouped into proliferating, mature and immature cell populations. These markers are specific to the mature adrenergic cells:
Wilcoxon ranksum test P-value for gene set overrepresentation: 8.02e-03
Mean rank of genes in gene set: 107
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| NTRK1 | 0.0032534 | 93 | GTEx | DepMap | Descartes | 0.58 | 261.82 |
| GAP43 | 0.0028822 | 121 | GTEx | DepMap | Descartes | 1.34 | 926.61 |
Adrenergic Fig 1D (Olsen)
Similar to above, but for selected adrenergic marker genes:
Wilcoxon ranksum test P-value for gene set overrepresentation: 8.35e-03
Mean rank of genes in gene set: 3662.18
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| NRG1 | 0.0098541 | 1 | GTEx | DepMap | Descartes | 14.28 | 1676.92 |
| ELAVL4 | 0.0034890 | 86 | GTEx | DepMap | Descartes | 2.69 | 866.51 |
| MAP1B | 0.0020545 | 210 | GTEx | DepMap | Descartes | 3.17 | 338.54 |
| STMN2 | 0.0012849 | 403 | GTEx | DepMap | Descartes | 4.48 | 3037.69 |
| RGS5 | 0.0009890 | 562 | GTEx | DepMap | Descartes | 1.20 | 266.08 |
| TH | 0.0009592 | 583 | GTEx | DepMap | Descartes | 1.19 | 819.63 |
| RTN1 | 0.0008397 | 665 | GTEx | DepMap | Descartes | 3.59 | 1305.32 |
| DBH | 0.0004944 | 1104 | GTEx | DepMap | Descartes | 0.89 | 410.35 |
| UCHL1 | -0.0009033 | 12081 | GTEx | DepMap | Descartes | 1.50 | 1162.83 |
| ISL1 | -0.0009593 | 12163 | GTEx | DepMap | Descartes | 0.67 | 341.76 |
| MLLT11 | -0.0013177 | 12426 | GTEx | DepMap | Descartes | 0.59 | 307.07 |
Sympathoblasts (Kildisiute)
Sympathoblasts markers obtained from Kildisiute et al, Supplmenentary Table 2, references supporting these genes are provided in Supp Table S2 of Kildisiute et al (PMID 33547074) https://www.science.org/doi/suppl/10.1126/sciadv.abd3311/suppl_file/abd3311_tables_s1_to_s12.xlsx:
Wilcoxon ranksum test P-value for gene set overrepresentation: 8.79e-03
Mean rank of genes in gene set: 1309.67
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| GAP43 | 0.0028822 | 121 | GTEx | DepMap | Descartes | 1.34 | 926.61 |
| BCL2 | 0.0007421 | 745 | GTEx | DepMap | Descartes | 2.43 | 408.62 |
| NPY | 0.0000625 | 3063 | GTEx | DepMap | Descartes | 6.96 | 12190.36 |
Below shows ranks on this GEP for literature curated gene lists for large gene sets
These include those reported as mesenchymal/adrenergic by Van Groningen et al.
High ranks indicate this gene is a driver of this GEP (note these results are not ordered).
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
VanGroningen Adrenergic Genes
Adrenergic marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.97e-01
Mean rank of genes in gene set: 6848.92
Median rank of genes in gene set: 7802
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CADM1 | 0.0071219 | 9 | GTEx | DepMap | Descartes | 6.18 | 941.18 |
| AUTS2 | 0.0064650 | 16 | GTEx | DepMap | Descartes | 17.39 | 2992.44 |
| ELAVL2 | 0.0061155 | 19 | GTEx | DepMap | Descartes | 2.84 | 961.56 |
| FAM155A | 0.0048086 | 45 | GTEx | DepMap | Descartes | 9.53 | 1365.94 |
| TMEM178B | 0.0039742 | 63 | GTEx | DepMap | Descartes | 3.40 | NA |
| MARCH11 | 0.0036818 | 75 | GTEx | DepMap | Descartes | 2.50 | NA |
| ELAVL4 | 0.0034890 | 86 | GTEx | DepMap | Descartes | 2.69 | 866.51 |
| GAP43 | 0.0028822 | 121 | GTEx | DepMap | Descartes | 1.34 | 926.61 |
| MAPT | 0.0028046 | 128 | GTEx | DepMap | Descartes | 1.60 | 300.47 |
| NRCAM | 0.0026228 | 143 | GTEx | DepMap | Descartes | 1.52 | 301.94 |
| NBEA | 0.0024167 | 166 | GTEx | DepMap | Descartes | 3.03 | 352.52 |
| RUNDC3B | 0.0021531 | 194 | GTEx | DepMap | Descartes | 0.92 | 297.80 |
| PPP1R9A | 0.0021446 | 195 | GTEx | DepMap | Descartes | 1.05 | 125.71 |
| MIAT | 0.0020780 | 203 | GTEx | DepMap | Descartes | 1.29 | 171.06 |
| MAP1B | 0.0020545 | 210 | GTEx | DepMap | Descartes | 3.17 | 338.54 |
| NNAT | 0.0020427 | 213 | GTEx | DepMap | Descartes | 1.51 | 1529.66 |
| NEFM | 0.0020036 | 219 | GTEx | DepMap | Descartes | 0.38 | 140.33 |
| UNC79 | 0.0016322 | 282 | GTEx | DepMap | Descartes | 1.31 | 186.39 |
| BMPR1B | 0.0015503 | 300 | GTEx | DepMap | Descartes | 3.93 | 895.44 |
| RBMS3 | 0.0015198 | 305 | GTEx | DepMap | Descartes | 5.30 | 794.16 |
| SCN3A | 0.0015075 | 310 | GTEx | DepMap | Descartes | 1.25 | 181.94 |
| TMOD1 | 0.0014106 | 341 | GTEx | DepMap | Descartes | 1.29 | 495.38 |
| CLASP2 | 0.0013728 | 357 | GTEx | DepMap | Descartes | 2.63 | 467.70 |
| ACTL6B | 0.0013579 | 367 | GTEx | DepMap | Descartes | 0.67 | 556.47 |
| CACNA1B | 0.0013508 | 370 | GTEx | DepMap | Descartes | 1.56 | 200.04 |
| NCOA7 | 0.0013411 | 373 | GTEx | DepMap | Descartes | 1.00 | NA |
| RNF165 | 0.0012951 | 398 | GTEx | DepMap | Descartes | 0.75 | 123.14 |
| STMN2 | 0.0012849 | 403 | GTEx | DepMap | Descartes | 4.48 | 3037.69 |
| GRB10 | 0.0012283 | 436 | GTEx | DepMap | Descartes | 0.27 | 63.41 |
| RNF144A | 0.0011527 | 476 | GTEx | DepMap | Descartes | 0.56 | 128.05 |
VanGroningen Mesenchymal Genes
Mesenchymal marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 8082.64
Median rank of genes in gene set: 9297
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ROBO1 | 0.0066089 | 14 | GTEx | DepMap | Descartes | 5.38 | 976.27 |
| DLC1 | 0.0058602 | 24 | GTEx | DepMap | Descartes | 8.85 | 1568.03 |
| ATP2B4 | 0.0031173 | 104 | GTEx | DepMap | Descartes | 0.74 | 111.01 |
| DMD | 0.0021943 | 186 | GTEx | DepMap | Descartes | 1.19 | 109.99 |
| ATXN1 | 0.0018402 | 237 | GTEx | DepMap | Descartes | 1.88 | 228.28 |
| NRP1 | 0.0015589 | 298 | GTEx | DepMap | Descartes | 0.86 | 187.28 |
| CD44 | 0.0014306 | 332 | GTEx | DepMap | Descartes | 1.08 | 259.52 |
| STAT3 | 0.0013664 | 362 | GTEx | DepMap | Descartes | 0.63 | 158.96 |
| EXT1 | 0.0013200 | 384 | GTEx | DepMap | Descartes | 1.54 | 237.03 |
| SCPEP1 | 0.0012435 | 427 | GTEx | DepMap | Descartes | 0.28 | 193.43 |
| PDE7B | 0.0011815 | 460 | GTEx | DepMap | Descartes | 0.19 | 45.19 |
| KIF13A | 0.0010937 | 511 | GTEx | DepMap | Descartes | 0.39 | 72.44 |
| LOXL2 | 0.0010409 | 537 | GTEx | DepMap | Descartes | 0.15 | 49.02 |
| PLAGL1 | 0.0007894 | 698 | GTEx | DepMap | Descartes | 0.31 | 65.88 |
| NR3C1 | 0.0007243 | 769 | GTEx | DepMap | Descartes | 0.25 | 44.36 |
| ADAM19 | 0.0007211 | 774 | GTEx | DepMap | Descartes | 0.19 | 36.97 |
| FILIP1L | 0.0007075 | 795 | GTEx | DepMap | Descartes | 0.54 | 154.51 |
| ACADVL | 0.0006833 | 822 | GTEx | DepMap | Descartes | 0.48 | 261.72 |
| CDH11 | 0.0006569 | 849 | GTEx | DepMap | Descartes | 0.19 | 34.32 |
| LPP | 0.0006111 | 908 | GTEx | DepMap | Descartes | 0.72 | 51.73 |
| RIN2 | 0.0005638 | 977 | GTEx | DepMap | Descartes | 0.15 | 43.22 |
| TMEFF2 | 0.0005522 | 999 | GTEx | DepMap | Descartes | 0.39 | 156.09 |
| RECK | 0.0005453 | 1011 | GTEx | DepMap | Descartes | 0.27 | 72.57 |
| NBR1 | 0.0005108 | 1061 | GTEx | DepMap | Descartes | 0.30 | 78.37 |
| AMMECR1 | 0.0005044 | 1076 | GTEx | DepMap | Descartes | 0.13 | 33.14 |
| SNAP23 | 0.0004985 | 1090 | GTEx | DepMap | Descartes | 0.17 | 84.39 |
| HIBADH | 0.0004958 | 1096 | GTEx | DepMap | Descartes | 0.43 | 286.79 |
| LIFR | 0.0004641 | 1153 | GTEx | DepMap | Descartes | 0.19 | 23.27 |
| ITM2C | 0.0004550 | 1176 | GTEx | DepMap | Descartes | 0.16 | 101.61 |
| PCSK5 | 0.0004482 | 1191 | GTEx | DepMap | Descartes | 0.17 | 21.31 |
Descartes adrenocortical markers
Top 50 marker genes of adrenocortical cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/adrenocortical/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 6.95e-01
Mean rank of genes in gene set: 6582.03
Median rank of genes in gene set: 7703
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| FRMD5 | 0.0036178 | 78 | GTEx | DepMap | Descartes | 2.13 | 547.97 |
| IGF1R | 0.0014864 | 317 | GTEx | DepMap | Descartes | 1.29 | 135.38 |
| CLU | 0.0013219 | 381 | GTEx | DepMap | Descartes | 0.68 | 311.31 |
| PEG3 | 0.0009656 | 580 | GTEx | DepMap | Descartes | 0.39 | NA |
| JAKMIP2 | 0.0007576 | 729 | GTEx | DepMap | Descartes | 0.67 | 93.31 |
| DNER | 0.0005915 | 939 | GTEx | DepMap | Descartes | 0.24 | 101.17 |
| NPC1 | 0.0004152 | 1256 | GTEx | DepMap | Descartes | 0.26 | 70.37 |
| HMGCS1 | 0.0003717 | 1363 | GTEx | DepMap | Descartes | 0.23 | 54.92 |
| SH3PXD2B | 0.0001627 | 2260 | GTEx | DepMap | Descartes | 0.14 | 22.95 |
| SLC1A2 | 0.0000636 | 3055 | GTEx | DepMap | Descartes | 0.31 | 32.98 |
| STAR | -0.0000478 | 4376 | GTEx | DepMap | Descartes | 0.00 | 0.73 |
| INHA | -0.0000794 | 4876 | GTEx | DepMap | Descartes | 0.01 | 8.93 |
| FREM2 | -0.0001636 | 6268 | GTEx | DepMap | Descartes | 0.01 | 0.41 |
| FDXR | -0.0002073 | 6973 | GTEx | DepMap | Descartes | 0.04 | 20.29 |
| FDPS | -0.0002144 | 7083 | GTEx | DepMap | Descartes | 0.32 | 194.34 |
| GSTA4 | -0.0002211 | 7194 | GTEx | DepMap | Descartes | 0.22 | 166.51 |
| GRAMD1B | -0.0002221 | 7212 | GTEx | DepMap | Descartes | 0.07 | 9.42 |
| CYB5B | -0.0002498 | 7623 | GTEx | DepMap | Descartes | 0.19 | 58.09 |
| HMGCR | -0.0002602 | 7783 | GTEx | DepMap | Descartes | 0.17 | 46.46 |
| FDX1 | -0.0002783 | 8026 | GTEx | DepMap | Descartes | 0.03 | 13.74 |
| DHCR24 | -0.0002861 | 8148 | GTEx | DepMap | Descartes | 0.08 | 18.69 |
| DHCR7 | -0.0002979 | 8335 | GTEx | DepMap | Descartes | 0.02 | 8.50 |
| POR | -0.0003019 | 8405 | GTEx | DepMap | Descartes | 0.10 | 51.83 |
| LDLR | -0.0003398 | 8909 | GTEx | DepMap | Descartes | 0.05 | 10.86 |
| SCAP | -0.0003500 | 9031 | GTEx | DepMap | Descartes | 0.16 | 46.92 |
| ERN1 | -0.0003795 | 9377 | GTEx | DepMap | Descartes | 0.04 | 5.34 |
| BAIAP2L1 | -0.0003805 | 9390 | GTEx | DepMap | Descartes | 0.02 | 5.95 |
| TM7SF2 | -0.0003879 | 9479 | GTEx | DepMap | Descartes | 0.08 | 46.62 |
| SGCZ | -0.0004021 | 9625 | GTEx | DepMap | Descartes | 0.31 | 45.58 |
| SLC16A9 | -0.0004099 | 9718 | GTEx | DepMap | Descartes | 0.10 | 28.34 |
Descartes chromaffin markers
Top 50 marker genes of chromaffin cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/chromaffin/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 7.14e-03
Mean rank of genes in gene set: 4891
Median rank of genes in gene set: 1918
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| RYR2 | 0.0082173 | 5 | GTEx | DepMap | Descartes | 7.52 | 588.12 |
| HS3ST5 | 0.0075159 | 8 | GTEx | DepMap | Descartes | 2.70 | 942.71 |
| ANKFN1 | 0.0064057 | 17 | GTEx | DepMap | Descartes | 1.36 | 440.69 |
| ELAVL2 | 0.0061155 | 19 | GTEx | DepMap | Descartes | 2.84 | 961.56 |
| IL7 | 0.0053210 | 31 | GTEx | DepMap | Descartes | 3.06 | 2006.66 |
| TMEM132C | 0.0037799 | 73 | GTEx | DepMap | Descartes | 3.81 | 1019.44 |
| MARCH11 | 0.0036818 | 75 | GTEx | DepMap | Descartes | 2.50 | NA |
| RBFOX1 | 0.0034736 | 88 | GTEx | DepMap | Descartes | 6.81 | 1790.47 |
| NTRK1 | 0.0032534 | 93 | GTEx | DepMap | Descartes | 0.58 | 261.82 |
| FAT3 | 0.0030731 | 106 | GTEx | DepMap | Descartes | 0.62 | 42.50 |
| KCNB2 | 0.0029505 | 118 | GTEx | DepMap | Descartes | 2.12 | 728.79 |
| GAP43 | 0.0028822 | 121 | GTEx | DepMap | Descartes | 1.34 | 926.61 |
| MAP1B | 0.0020545 | 210 | GTEx | DepMap | Descartes | 3.17 | 338.54 |
| CNKSR2 | 0.0016523 | 276 | GTEx | DepMap | Descartes | 0.97 | 155.21 |
| STMN2 | 0.0012849 | 403 | GTEx | DepMap | Descartes | 4.48 | 3037.69 |
| TUBB2A | 0.0009357 | 596 | GTEx | DepMap | Descartes | 0.89 | 684.64 |
| PLXNA4 | 0.0009189 | 611 | GTEx | DepMap | Descartes | 0.48 | 45.25 |
| REEP1 | 0.0007210 | 775 | GTEx | DepMap | Descartes | 0.40 | 126.70 |
| CNTFR | 0.0006768 | 832 | GTEx | DepMap | Descartes | 0.28 | 184.16 |
| TMEFF2 | 0.0005522 | 999 | GTEx | DepMap | Descartes | 0.39 | 156.09 |
| MAB21L1 | 0.0002234 | 1918 | GTEx | DepMap | Descartes | 0.22 | 93.95 |
| NPY | 0.0000625 | 3063 | GTEx | DepMap | Descartes | 6.96 | 12190.36 |
| TUBB2B | 0.0000543 | 3138 | GTEx | DepMap | Descartes | 2.44 | 1573.80 |
| BASP1 | -0.0000036 | 3772 | GTEx | DepMap | Descartes | 1.05 | 740.86 |
| RPH3A | -0.0000346 | 4188 | GTEx | DepMap | Descartes | 0.04 | 8.87 |
| EPHA6 | -0.0000435 | 4313 | GTEx | DepMap | Descartes | 0.86 | 258.20 |
| GREM1 | -0.0002851 | 8135 | GTEx | DepMap | Descartes | 0.00 | 0.48 |
| RGMB | -0.0003749 | 9331 | GTEx | DepMap | Descartes | 0.35 | 100.71 |
| PTCHD1 | -0.0004925 | 10462 | GTEx | DepMap | Descartes | 0.12 | 11.01 |
| TUBA1A | -0.0006068 | 11211 | GTEx | DepMap | Descartes | 4.49 | 2987.08 |
Descartes Vascular_endothelial markers
Top 50 marker genes of Vascular_endothelial cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/vascular_endothelial/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 8.37e-01
Mean rank of genes in gene set: 6851.47
Median rank of genes in gene set: 6984.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CDH13 | 0.0023649 | 169 | GTEx | DepMap | Descartes | 0.47 | 80.82 |
| EFNB2 | 0.0017053 | 264 | GTEx | DepMap | Descartes | 0.42 | 108.37 |
| CEACAM1 | 0.0006405 | 868 | GTEx | DepMap | Descartes | 0.08 | 30.90 |
| TEK | 0.0005756 | 962 | GTEx | DepMap | Descartes | 0.05 | 14.29 |
| IRX3 | 0.0004247 | 1231 | GTEx | DepMap | Descartes | 0.01 | 4.60 |
| EHD3 | 0.0002947 | 1611 | GTEx | DepMap | Descartes | 0.03 | 9.69 |
| GALNT15 | 0.0001877 | 2093 | GTEx | DepMap | Descartes | 0.01 | NA |
| F8 | -0.0000644 | 4619 | GTEx | DepMap | Descartes | 0.00 | 0.24 |
| CYP26B1 | -0.0000694 | 4697 | GTEx | DepMap | Descartes | 0.01 | 1.57 |
| CRHBP | -0.0001279 | 5698 | GTEx | DepMap | Descartes | 0.00 | 2.28 |
| TMEM88 | -0.0001638 | 6272 | GTEx | DepMap | Descartes | 0.05 | 80.56 |
| SHE | -0.0001667 | 6324 | GTEx | DepMap | Descartes | 0.01 | 0.84 |
| NOTCH4 | -0.0001850 | 6599 | GTEx | DepMap | Descartes | 0.03 | 3.84 |
| ROBO4 | -0.0001856 | 6609 | GTEx | DepMap | Descartes | 0.01 | 1.80 |
| KANK3 | -0.0001934 | 6733 | GTEx | DepMap | Descartes | 0.01 | 3.58 |
| TIE1 | -0.0001977 | 6808 | GTEx | DepMap | Descartes | 0.01 | 2.00 |
| BTNL9 | -0.0002005 | 6858 | GTEx | DepMap | Descartes | 0.00 | 1.04 |
| SLCO2A1 | -0.0002023 | 6884 | GTEx | DepMap | Descartes | 0.01 | 2.23 |
| RASIP1 | -0.0002073 | 6970 | GTEx | DepMap | Descartes | 0.01 | 3.00 |
| MMRN2 | -0.0002090 | 6999 | GTEx | DepMap | Descartes | 0.01 | 1.44 |
| CLDN5 | -0.0002101 | 7017 | GTEx | DepMap | Descartes | 0.01 | 4.65 |
| NPR1 | -0.0002271 | 7282 | GTEx | DepMap | Descartes | 0.00 | 0.04 |
| KDR | -0.0002323 | 7371 | GTEx | DepMap | Descartes | 0.00 | 0.46 |
| CDH5 | -0.0002324 | 7373 | GTEx | DepMap | Descartes | 0.01 | 0.99 |
| ESM1 | -0.0002425 | 7527 | GTEx | DepMap | Descartes | 0.00 | 1.06 |
| CALCRL | -0.0002738 | 7976 | GTEx | DepMap | Descartes | 0.03 | 5.09 |
| HYAL2 | -0.0003149 | 8585 | GTEx | DepMap | Descartes | 0.04 | 12.94 |
| FLT4 | -0.0003432 | 8948 | GTEx | DepMap | Descartes | 0.00 | 0.20 |
| SHANK3 | -0.0003712 | 9290 | GTEx | DepMap | Descartes | 0.02 | 3.68 |
| MYRIP | -0.0004149 | 9765 | GTEx | DepMap | Descartes | 0.17 | 35.25 |
Descartes stromal markers
Top 50 marker genes of stromal cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/stromal/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 8417.34
Median rank of genes in gene set: 9840.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| FREM1 | 0.0015929 | 293 | GTEx | DepMap | Descartes | 0.32 | 40.31 |
| PRICKLE1 | 0.0009127 | 618 | GTEx | DepMap | Descartes | 0.51 | 117.59 |
| CDH11 | 0.0006569 | 849 | GTEx | DepMap | Descartes | 0.19 | 34.32 |
| PAMR1 | 0.0002945 | 1613 | GTEx | DepMap | Descartes | 0.07 | 29.96 |
| LRRC17 | 0.0002368 | 1859 | GTEx | DepMap | Descartes | 0.05 | 30.28 |
| SCARA5 | 0.0000566 | 3119 | GTEx | DepMap | Descartes | 0.00 | 1.10 |
| OGN | 0.0000337 | 3348 | GTEx | DepMap | Descartes | 0.04 | 11.99 |
| ABCA6 | 0.0000029 | 3695 | GTEx | DepMap | Descartes | 0.03 | 5.19 |
| GAS2 | -0.0000527 | 4448 | GTEx | DepMap | Descartes | 0.10 | 55.06 |
| ELN | -0.0001203 | 5554 | GTEx | DepMap | Descartes | 0.06 | 17.84 |
| LAMC3 | -0.0001484 | 6019 | GTEx | DepMap | Descartes | 0.01 | 1.58 |
| PCDH18 | -0.0001521 | 6066 | GTEx | DepMap | Descartes | 0.00 | 0.16 |
| ADAMTS2 | -0.0001606 | 6213 | GTEx | DepMap | Descartes | 0.08 | 13.93 |
| RSPO3 | -0.0002126 | 7051 | GTEx | DepMap | Descartes | 0.00 | NA |
| ADAMTSL3 | -0.0002929 | 8268 | GTEx | DepMap | Descartes | 0.03 | 4.00 |
| ABCC9 | -0.0003191 | 8639 | GTEx | DepMap | Descartes | 0.00 | 0.57 |
| CLDN11 | -0.0003420 | 8934 | GTEx | DepMap | Descartes | 0.12 | 47.80 |
| LOX | -0.0003518 | 9053 | GTEx | DepMap | Descartes | 0.00 | 0.37 |
| GLI2 | -0.0003641 | 9214 | GTEx | DepMap | Descartes | 0.01 | 1.03 |
| ITGA11 | -0.0003830 | 9423 | GTEx | DepMap | Descartes | 0.00 | 0.32 |
| SFRP2 | -0.0003939 | 9530 | GTEx | DepMap | Descartes | 0.00 | 0.26 |
| EDNRA | -0.0004212 | 9824 | GTEx | DepMap | Descartes | 0.01 | 2.66 |
| CD248 | -0.0004245 | 9857 | GTEx | DepMap | Descartes | 0.00 | 0.72 |
| DKK2 | -0.0004263 | 9872 | GTEx | DepMap | Descartes | 0.02 | 5.13 |
| PDGFRA | -0.0004774 | 10348 | GTEx | DepMap | Descartes | 0.00 | 0.21 |
| COL27A1 | -0.0005035 | 10542 | GTEx | DepMap | Descartes | 0.01 | 1.00 |
| COL12A1 | -0.0005047 | 10553 | GTEx | DepMap | Descartes | 0.01 | 0.81 |
| IGFBP3 | -0.0005135 | 10627 | GTEx | DepMap | Descartes | 0.01 | 3.02 |
| PRRX1 | -0.0005245 | 10703 | GTEx | DepMap | Descartes | 0.00 | 1.10 |
| CCDC80 | -0.0005377 | 10789 | GTEx | DepMap | Descartes | 0.01 | 0.63 |
Descartes sympathoblasts markers
Top 50 marker genes of sympathoblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/sympathoblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 4.09e-03
Mean rank of genes in gene set: 4723.32
Median rank of genes in gene set: 3031.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ROBO1 | 0.0066089 | 14 | GTEx | DepMap | Descartes | 5.38 | 976.27 |
| AGBL4 | 0.0057700 | 25 | GTEx | DepMap | Descartes | 2.85 | 941.23 |
| FAM155A | 0.0048086 | 45 | GTEx | DepMap | Descartes | 9.53 | 1365.94 |
| GRID2 | 0.0046215 | 48 | GTEx | DepMap | Descartes | 1.54 | 373.20 |
| NTNG1 | 0.0039028 | 66 | GTEx | DepMap | Descartes | 1.57 | 428.02 |
| CDH12 | 0.0036341 | 77 | GTEx | DepMap | Descartes | 0.38 | 116.20 |
| KCTD16 | 0.0034751 | 87 | GTEx | DepMap | Descartes | 1.86 | 171.30 |
| PACRG | 0.0029522 | 117 | GTEx | DepMap | Descartes | 0.78 | 629.21 |
| TENM1 | 0.0026767 | 139 | GTEx | DepMap | Descartes | 0.83 | NA |
| KSR2 | 0.0025181 | 154 | GTEx | DepMap | Descartes | 0.71 | 50.28 |
| SLC18A1 | 0.0018360 | 238 | GTEx | DepMap | Descartes | 0.37 | 172.91 |
| MGAT4C | 0.0017556 | 251 | GTEx | DepMap | Descartes | 0.60 | 31.10 |
| SLC24A2 | 0.0013853 | 353 | GTEx | DepMap | Descartes | 0.11 | 13.74 |
| CNTN3 | 0.0011313 | 494 | GTEx | DepMap | Descartes | 0.10 | 24.19 |
| UNC80 | 0.0010412 | 536 | GTEx | DepMap | Descartes | 1.02 | 94.97 |
| GCH1 | 0.0007236 | 771 | GTEx | DepMap | Descartes | 0.42 | 192.30 |
| PCSK2 | 0.0006538 | 855 | GTEx | DepMap | Descartes | 0.32 | 92.65 |
| TBX20 | 0.0005947 | 932 | GTEx | DepMap | Descartes | 0.19 | 135.17 |
| SLC35F3 | 0.0001027 | 2665 | GTEx | DepMap | Descartes | 0.20 | 83.26 |
| PCSK1N | 0.0000291 | 3398 | GTEx | DepMap | Descartes | 1.60 | 1905.61 |
| CHGB | 0.0000277 | 3405 | GTEx | DepMap | Descartes | 1.45 | 771.96 |
| GRM7 | 0.0000019 | 3709 | GTEx | DepMap | Descartes | 0.35 | 107.20 |
| LAMA3 | -0.0000501 | 4405 | GTEx | DepMap | Descartes | 0.09 | 10.45 |
| TIAM1 | -0.0000640 | 4616 | GTEx | DepMap | Descartes | 0.66 | 114.10 |
| SORCS3 | -0.0001632 | 6259 | GTEx | DepMap | Descartes | 0.02 | 3.77 |
| DGKK | -0.0001986 | 6822 | GTEx | DepMap | Descartes | 0.01 | 1.08 |
| PENK | -0.0002888 | 8194 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| C1QL1 | -0.0004740 | 10310 | GTEx | DepMap | Descartes | 0.07 | 62.79 |
| SPOCK3 | -0.0006082 | 11223 | GTEx | DepMap | Descartes | 0.19 | 85.30 |
| GALNTL6 | -0.0007266 | 11687 | GTEx | DepMap | Descartes | 0.39 | 119.66 |
Descartes erythroblasts markers
Top 50 marker genes of erythroblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/erythroblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.24e-02
Mean rank of genes in gene set: 4766.14
Median rank of genes in gene set: 4136
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| SOX6 | 0.0060181 | 22 | GTEx | DepMap | Descartes | 1.65 | 246.18 |
| EPB41 | 0.0020108 | 218 | GTEx | DepMap | Descartes | 0.78 | 155.81 |
| TFR2 | 0.0009320 | 598 | GTEx | DepMap | Descartes | 0.33 | 134.98 |
| SNCA | 0.0009281 | 602 | GTEx | DepMap | Descartes | 0.23 | 83.28 |
| ANK1 | 0.0006565 | 851 | GTEx | DepMap | Descartes | 0.25 | 37.40 |
| DENND4A | 0.0005903 | 942 | GTEx | DepMap | Descartes | 0.73 | 107.92 |
| ALAS2 | 0.0001958 | 2046 | GTEx | DepMap | Descartes | 0.00 | 0.14 |
| TRAK2 | 0.0001854 | 2108 | GTEx | DepMap | Descartes | 0.15 | 29.47 |
| MARCH3 | 0.0001327 | 2453 | GTEx | DepMap | Descartes | 0.18 | NA |
| RGS6 | 0.0001183 | 2554 | GTEx | DepMap | Descartes | 0.02 | 5.22 |
| SLC25A37 | 0.0000970 | 2720 | GTEx | DepMap | Descartes | 0.11 | 30.08 |
| RHD | 0.0000961 | 2732 | GTEx | DepMap | Descartes | 0.03 | 12.35 |
| XPO7 | 0.0000012 | 3714 | GTEx | DepMap | Descartes | 0.25 | 60.88 |
| TMCC2 | -0.0000078 | 3826 | GTEx | DepMap | Descartes | 0.04 | 11.39 |
| SPTB | -0.0000309 | 4136 | GTEx | DepMap | Descartes | 0.07 | 7.76 |
| SLC25A21 | -0.0000368 | 4222 | GTEx | DepMap | Descartes | 0.01 | 2.57 |
| CAT | -0.0000468 | 4361 | GTEx | DepMap | Descartes | 0.07 | 41.44 |
| FECH | -0.0001384 | 5854 | GTEx | DepMap | Descartes | 0.07 | 11.98 |
| SLC4A1 | -0.0001604 | 6205 | GTEx | DepMap | Descartes | 0.00 | 0.44 |
| TSPAN5 | -0.0001639 | 6276 | GTEx | DepMap | Descartes | 0.66 | 197.84 |
| GYPC | -0.0002181 | 7141 | GTEx | DepMap | Descartes | 0.01 | 4.22 |
| BLVRB | -0.0002631 | 7822 | GTEx | DepMap | Descartes | 0.03 | 24.27 |
| CPOX | -0.0002869 | 8159 | GTEx | DepMap | Descartes | 0.03 | 13.22 |
| ABCB10 | -0.0002963 | 8311 | GTEx | DepMap | Descartes | 0.09 | 28.08 |
| SELENBP1 | -0.0003081 | 8480 | GTEx | DepMap | Descartes | 0.00 | 0.20 |
| MICAL2 | -0.0004254 | 9861 | GTEx | DepMap | Descartes | 0.02 | 3.11 |
| SPECC1 | -0.0004462 | 10067 | GTEx | DepMap | Descartes | 0.02 | 2.00 |
| RAPGEF2 | -0.0004721 | 10300 | GTEx | DepMap | Descartes | 0.47 | 70.98 |
| GCLC | -0.0007107 | 11637 | GTEx | DepMap | Descartes | 0.08 | 25.67 |
| NA | NA | NA | NA | NA | NA | NA | NA |
Descartes myeloid markers
Top 50 marker genes of myeloid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/myeloid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 10160.45
Median rank of genes in gene set: 11216.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| FMN1 | 0.0016476 | 279 | GTEx | DepMap | Descartes | 1.88 | 184.09 |
| CD163L1 | 0.0001994 | 2029 | GTEx | DepMap | Descartes | 0.27 | 75.57 |
| HRH1 | -0.0000512 | 4425 | GTEx | DepMap | Descartes | 0.04 | 11.66 |
| WWP1 | -0.0001787 | 6501 | GTEx | DepMap | Descartes | 0.18 | 45.05 |
| SFMBT2 | -0.0002418 | 7512 | GTEx | DepMap | Descartes | 0.18 | 28.53 |
| FGL2 | -0.0002531 | 7664 | GTEx | DepMap | Descartes | 0.01 | 2.47 |
| CPVL | -0.0002735 | 7969 | GTEx | DepMap | Descartes | 0.04 | 24.37 |
| CTSS | -0.0003009 | 8384 | GTEx | DepMap | Descartes | 0.05 | 15.47 |
| ATP8B4 | -0.0003337 | 8822 | GTEx | DepMap | Descartes | 0.03 | 7.00 |
| MS4A4A | -0.0003734 | 9313 | GTEx | DepMap | Descartes | 0.01 | 5.18 |
| HCK | -0.0004329 | 9935 | GTEx | DepMap | Descartes | 0.01 | 7.52 |
| CYBB | -0.0004518 | 10118 | GTEx | DepMap | Descartes | 0.01 | 2.42 |
| FGD2 | -0.0004678 | 10273 | GTEx | DepMap | Descartes | 0.01 | 2.20 |
| CSF1R | -0.0005014 | 10530 | GTEx | DepMap | Descartes | 0.01 | 3.33 |
| CD163 | -0.0005110 | 10610 | GTEx | DepMap | Descartes | 0.00 | 0.78 |
| AXL | -0.0005408 | 10807 | GTEx | DepMap | Descartes | 0.02 | 5.19 |
| ADAP2 | -0.0005525 | 10881 | GTEx | DepMap | Descartes | 0.04 | 15.34 |
| CD14 | -0.0005825 | 11081 | GTEx | DepMap | Descartes | 0.02 | 14.22 |
| SPP1 | -0.0006063 | 11208 | GTEx | DepMap | Descartes | 0.35 | 316.06 |
| IFNGR1 | -0.0006084 | 11225 | GTEx | DepMap | Descartes | 0.07 | 36.12 |
| CTSC | -0.0006571 | 11443 | GTEx | DepMap | Descartes | 0.02 | 3.68 |
| SLCO2B1 | -0.0006782 | 11512 | GTEx | DepMap | Descartes | 0.02 | 3.43 |
| SLC1A3 | -0.0007306 | 11701 | GTEx | DepMap | Descartes | 0.01 | 2.89 |
| PTPRE | -0.0007492 | 11745 | GTEx | DepMap | Descartes | 0.05 | 8.18 |
| LGMN | -0.0007649 | 11784 | GTEx | DepMap | Descartes | 0.06 | 34.16 |
| MERTK | -0.0007759 | 11822 | GTEx | DepMap | Descartes | 0.01 | 2.83 |
| ITPR2 | -0.0008440 | 11975 | GTEx | DepMap | Descartes | 0.13 | 12.31 |
| RBPJ | -0.0008545 | 12001 | GTEx | DepMap | Descartes | 0.48 | 99.30 |
| TGFBI | -0.0008993 | 12070 | GTEx | DepMap | Descartes | 0.02 | 3.44 |
| MSR1 | -0.0009494 | 12152 | GTEx | DepMap | Descartes | 0.02 | 5.03 |
Descartes Schwann markers
Top 50 marker genes of Schwann cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/schwann/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.06e-01
Mean rank of genes in gene set: 5827.02
Median rank of genes in gene set: 5468
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| LRRTM4 | 0.0090314 | 4 | GTEx | DepMap | Descartes | 6.03 | 2347.41 |
| NRXN1 | 0.0044759 | 51 | GTEx | DepMap | Descartes | 3.48 | 476.71 |
| PPP2R2B | 0.0026986 | 137 | GTEx | DepMap | Descartes | 1.51 | 180.78 |
| DST | 0.0021884 | 189 | GTEx | DepMap | Descartes | 3.05 | 176.15 |
| SORCS1 | 0.0020137 | 217 | GTEx | DepMap | Descartes | 1.81 | 325.43 |
| TRPM3 | 0.0018513 | 235 | GTEx | DepMap | Descartes | 0.33 | 39.22 |
| SOX5 | 0.0016890 | 270 | GTEx | DepMap | Descartes | 0.99 | 183.10 |
| LAMA4 | 0.0016193 | 286 | GTEx | DepMap | Descartes | 0.15 | 26.35 |
| SCN7A | 0.0013697 | 360 | GTEx | DepMap | Descartes | 0.42 | 74.41 |
| GFRA3 | 0.0012402 | 428 | GTEx | DepMap | Descartes | 0.26 | 167.56 |
| XKR4 | 0.0011964 | 447 | GTEx | DepMap | Descartes | 0.37 | 23.13 |
| PMP22 | 0.0011673 | 470 | GTEx | DepMap | Descartes | 0.55 | 389.20 |
| IL1RAPL1 | 0.0008760 | 641 | GTEx | DepMap | Descartes | 0.35 | 115.75 |
| PAG1 | 0.0006882 | 815 | GTEx | DepMap | Descartes | 0.74 | 82.91 |
| NRXN3 | 0.0006316 | 877 | GTEx | DepMap | Descartes | 0.36 | 56.64 |
| COL25A1 | 0.0005265 | 1039 | GTEx | DepMap | Descartes | 0.06 | 9.06 |
| ADAMTS5 | 0.0002600 | 1764 | GTEx | DepMap | Descartes | 0.02 | 2.53 |
| SLC35F1 | 0.0000737 | 2954 | GTEx | DepMap | Descartes | 0.08 | 17.96 |
| PTN | -0.0000155 | 3910 | GTEx | DepMap | Descartes | 0.27 | 203.45 |
| EGFLAM | -0.0000613 | 4579 | GTEx | DepMap | Descartes | 0.15 | 37.77 |
| GRIK3 | -0.0000999 | 5219 | GTEx | DepMap | Descartes | 0.04 | 4.43 |
| HMGA2 | -0.0001120 | 5432 | GTEx | DepMap | Descartes | 0.00 | 0.81 |
| MDGA2 | -0.0001168 | 5504 | GTEx | DepMap | Descartes | 0.00 | 0.86 |
| PLP1 | -0.0001207 | 5564 | GTEx | DepMap | Descartes | 0.00 | 1.17 |
| MPZ | -0.0001634 | 6265 | GTEx | DepMap | Descartes | 0.01 | 5.64 |
| ERBB3 | -0.0002216 | 7206 | GTEx | DepMap | Descartes | 0.01 | 2.08 |
| EDNRB | -0.0002822 | 8096 | GTEx | DepMap | Descartes | 0.00 | 0.50 |
| OLFML2A | -0.0003469 | 8993 | GTEx | DepMap | Descartes | 0.00 | 0.37 |
| PTPRZ1 | -0.0003604 | 9161 | GTEx | DepMap | Descartes | 0.01 | 0.87 |
| SFRP1 | -0.0005050 | 10559 | GTEx | DepMap | Descartes | 0.06 | 15.00 |
Descartes Megakaryocytes markers
Top 50 marker genes of Megakaryocytes cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/megakaryocytes/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.73e-01
Mean rank of genes in gene set: 7316.49
Median rank of genes in gene set: 7012
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TMSB4X | 0.0027689 | 134 | GTEx | DepMap | Descartes | 6.00 | 4689.67 |
| INPP4B | 0.0025878 | 147 | GTEx | DepMap | Descartes | 1.07 | 162.41 |
| RAB27B | 0.0011492 | 480 | GTEx | DepMap | Descartes | 0.18 | 33.11 |
| STON2 | 0.0011368 | 488 | GTEx | DepMap | Descartes | 0.18 | 53.34 |
| DOK6 | 0.0008042 | 687 | GTEx | DepMap | Descartes | 0.39 | 56.56 |
| ITGA2B | 0.0007046 | 800 | GTEx | DepMap | Descartes | 0.11 | 41.59 |
| CD9 | 0.0003739 | 1355 | GTEx | DepMap | Descartes | 0.23 | 158.69 |
| TUBB1 | 0.0001424 | 2383 | GTEx | DepMap | Descartes | 0.00 | 0.92 |
| TLN1 | 0.0000874 | 2811 | GTEx | DepMap | Descartes | 0.11 | 16.95 |
| TGFB1 | -0.0000418 | 4295 | GTEx | DepMap | Descartes | 0.13 | 63.16 |
| PRKAR2B | -0.0000550 | 4484 | GTEx | DepMap | Descartes | 0.40 | 142.40 |
| P2RX1 | -0.0000653 | 4631 | GTEx | DepMap | Descartes | 0.00 | 2.30 |
| SLC24A3 | -0.0001160 | 5490 | GTEx | DepMap | Descartes | 0.07 | 19.17 |
| MYLK | -0.0001288 | 5711 | GTEx | DepMap | Descartes | 0.03 | 3.88 |
| ACTN1 | -0.0001306 | 5743 | GTEx | DepMap | Descartes | 0.29 | 76.73 |
| ITGB3 | -0.0001370 | 5833 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MMRN1 | -0.0001399 | 5876 | GTEx | DepMap | Descartes | 0.00 | 0.60 |
| GP1BA | -0.0001405 | 5894 | GTEx | DepMap | Descartes | 0.02 | 7.87 |
| RAP1B | -0.0001578 | 6162 | GTEx | DepMap | Descartes | 0.20 | 19.65 |
| PSTPIP2 | -0.0001772 | 6471 | GTEx | DepMap | Descartes | 0.02 | 8.67 |
| ARHGAP6 | -0.0001878 | 6650 | GTEx | DepMap | Descartes | 0.02 | 3.16 |
| ANGPT1 | -0.0002029 | 6892 | GTEx | DepMap | Descartes | 0.01 | 3.10 |
| UBASH3B | -0.0002097 | 7012 | GTEx | DepMap | Descartes | 0.07 | 12.36 |
| TRPC6 | -0.0002228 | 7224 | GTEx | DepMap | Descartes | 0.01 | 1.47 |
| VCL | -0.0002742 | 7980 | GTEx | DepMap | Descartes | 0.11 | 17.04 |
| FERMT3 | -0.0002959 | 8304 | GTEx | DepMap | Descartes | 0.02 | 7.85 |
| SPN | -0.0003088 | 8494 | GTEx | DepMap | Descartes | 0.01 | 1.36 |
| PLEK | -0.0004257 | 9864 | GTEx | DepMap | Descartes | 0.01 | 3.93 |
| CD84 | -0.0004650 | 10251 | GTEx | DepMap | Descartes | 0.01 | 1.62 |
| LTBP1 | -0.0004763 | 10339 | GTEx | DepMap | Descartes | 0.02 | 4.09 |
Descartes Lyphoid markers
Top 50 marker genes of Lyphoid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/lymphoid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 4.02e-01
Mean rank of genes in gene set: 6136.21
Median rank of genes in gene set: 6097.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| RAP1GAP2 | 0.0035586 | 82 | GTEx | DepMap | Descartes | 2.06 | 406.87 |
| STK39 | 0.0026238 | 142 | GTEx | DepMap | Descartes | 1.29 | 515.49 |
| NCALD | 0.0014413 | 329 | GTEx | DepMap | Descartes | 0.39 | 132.42 |
| CD44 | 0.0014306 | 332 | GTEx | DepMap | Descartes | 1.08 | 259.52 |
| FYN | 0.0012662 | 417 | GTEx | DepMap | Descartes | 1.40 | 492.47 |
| FOXP1 | 0.0011448 | 484 | GTEx | DepMap | Descartes | 1.83 | 252.43 |
| SORL1 | 0.0011353 | 490 | GTEx | DepMap | Descartes | 0.40 | 46.13 |
| TOX | 0.0009243 | 607 | GTEx | DepMap | Descartes | 1.37 | 438.94 |
| SCML4 | 0.0009108 | 620 | GTEx | DepMap | Descartes | 0.31 | 86.89 |
| PITPNC1 | 0.0008461 | 661 | GTEx | DepMap | Descartes | 1.03 | 200.46 |
| CELF2 | 0.0008239 | 675 | GTEx | DepMap | Descartes | 0.35 | 56.17 |
| BCL2 | 0.0007421 | 745 | GTEx | DepMap | Descartes | 2.43 | 408.62 |
| MCTP2 | 0.0004317 | 1217 | GTEx | DepMap | Descartes | 0.06 | 9.05 |
| GNG2 | 0.0002309 | 1883 | GTEx | DepMap | Descartes | 0.65 | 223.15 |
| ABLIM1 | 0.0000781 | 2897 | GTEx | DepMap | Descartes | 0.25 | 39.95 |
| CCL5 | 0.0000460 | 3208 | GTEx | DepMap | Descartes | 0.02 | 21.18 |
| LCP1 | -0.0000985 | 5183 | GTEx | DepMap | Descartes | 0.04 | 12.15 |
| BACH2 | -0.0001064 | 5346 | GTEx | DepMap | Descartes | 0.70 | 97.82 |
| LEF1 | -0.0001211 | 5575 | GTEx | DepMap | Descartes | 0.02 | 7.74 |
| DOCK10 | -0.0001249 | 5641 | GTEx | DepMap | Descartes | 0.45 | 73.93 |
| EVL | -0.0001255 | 5652 | GTEx | DepMap | Descartes | 1.11 | 372.47 |
| SKAP1 | -0.0001816 | 6543 | GTEx | DepMap | Descartes | 0.06 | 40.43 |
| SAMD3 | -0.0001955 | 6767 | GTEx | DepMap | Descartes | 0.02 | 6.15 |
| ARHGDIB | -0.0001999 | 6838 | GTEx | DepMap | Descartes | 0.01 | 11.71 |
| PDE3B | -0.0003077 | 8477 | GTEx | DepMap | Descartes | 0.36 | 74.56 |
| ANKRD44 | -0.0003445 | 8965 | GTEx | DepMap | Descartes | 0.31 | 53.24 |
| RCSD1 | -0.0004062 | 9678 | GTEx | DepMap | Descartes | 0.02 | 3.38 |
| ETS1 | -0.0004310 | 9912 | GTEx | DepMap | Descartes | 0.03 | 7.45 |
| PRKCH | -0.0004351 | 9957 | GTEx | DepMap | Descartes | 0.05 | 14.06 |
| PTPRC | -0.0004660 | 10262 | GTEx | DepMap | Descartes | 0.04 | 9.26 |
Below shows the ranks on this GEP for immune marker gene lists curated by CellTypist (https://www.celltypist.org/encyclopedia).
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
HSC/MPP: MEMP (curated markers)
shared progenitors which are derived from common myeloid progenitors and will differentiate into megakaryocytes:
Wilcoxon ranksum test P-value for gene set overrepresentation: 4.17e-02
Mean rank of genes in gene set: 1840
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ITGA2B | 0.0007046 | 800 | GTEx | DepMap | Descartes | 0.11 | 41.59 |
| GATA2 | 0.0000794 | 2880 | GTEx | DepMap | Descartes | 0.42 | 168.69 |
Megakaryocyte precursor: Megakaryocyte precursor (curated markers)
megakaryocyte precursors in the bone marrow that are committed to megakaryocytes:
Wilcoxon ranksum test P-value for gene set overrepresentation: 5.77e-02
Mean rank of genes in gene set: 2242.5
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| PRSS57 | 0.0002962 | 1605 | GTEx | DepMap | Descartes | 0.01 | 8.22 |
| GATA2 | 0.0000794 | 2880 | GTEx | DepMap | Descartes | 0.42 | 168.69 |
B-cell lineage: Large pre-B cells (model markers)
proliferative B lymphocyte precursors derived from Pro-B cells and expressing membrane μ chains with surrogate light chains in their receptors:
Wilcoxon ranksum test P-value for gene set overrepresentation: 6.00e-02
Mean rank of genes in gene set: 641
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| IL1RAPL1 | 0.000876 | 641 | GTEx | DepMap | Descartes | 0.35 | 115.75 |