Program: 5. Neuroblastoma: Adrenergic I (pre-chromaffin).
Submit a comment on this gene expression program’s interpretation: CLICK
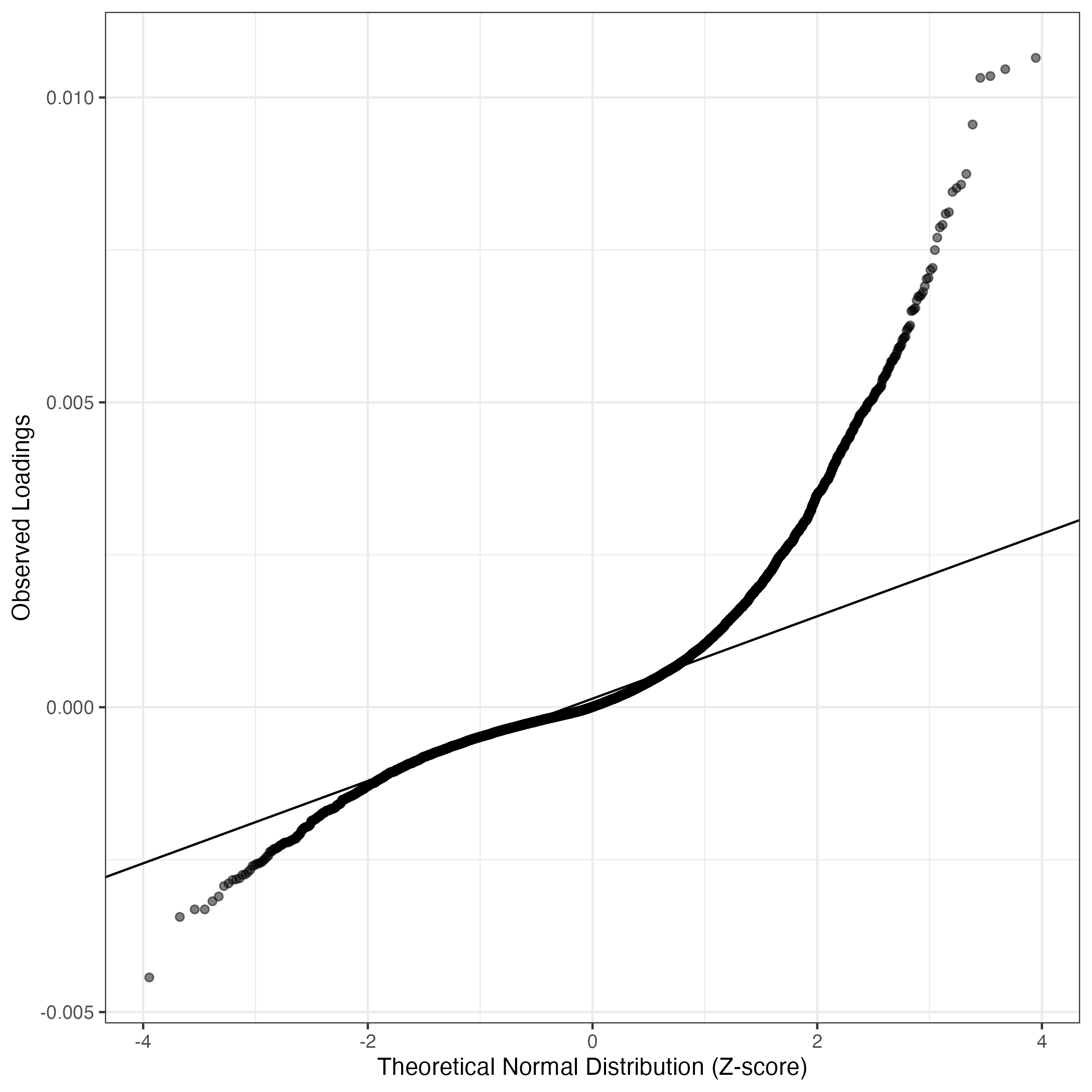
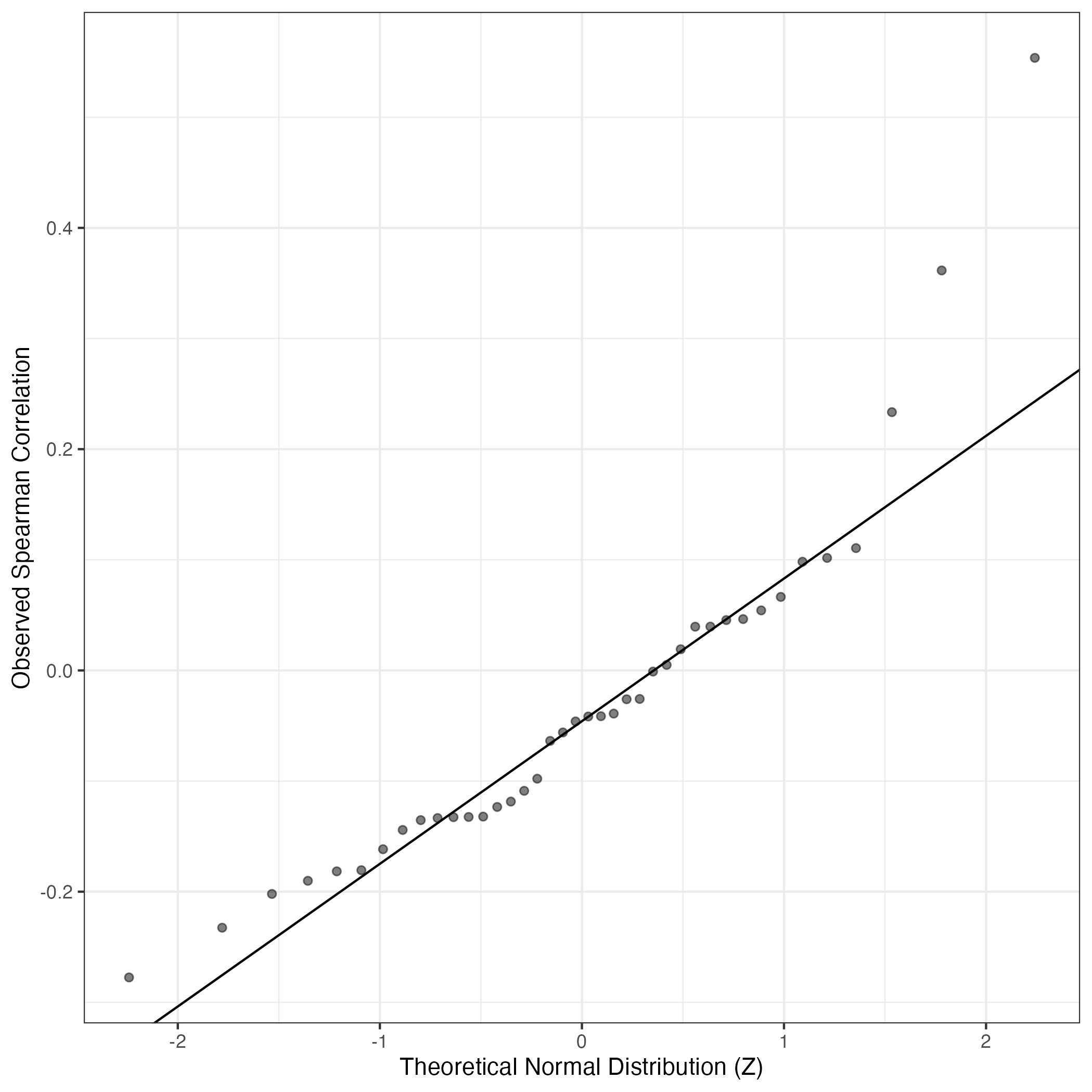
QQ-plot of gene loadings, averaged over both independent splits of the data
This plot highlights the relative contribution of each gene to the GEP
Top genes driving this program.
Note: Decartes website is buggy, try refreshing. Also, Decartes fetal adrenal data have been collected at specific time points (89-122 days), all possible cell types of interest may not be represented, do not overinterpret.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
| Gene | Loading | Gene.Name | GTEx | DepMap | Descartes | Mean.Counts | Mean.Tpm | |
|---|---|---|---|---|---|---|---|---|
| 1 | RGS5 | 0.0106499 | regulator of G protein signaling 5 | GTEx | DepMap | Descartes | 17.67 | 940.52 |
| 2 | NTRK1 | 0.0104639 | neurotrophic receptor tyrosine kinase 1 | GTEx | DepMap | Descartes | 4.14 | 465.49 |
| 3 | PKIB | 0.0103512 | cAMP-dependent protein kinase inhibitor beta | GTEx | DepMap | Descartes | 6.00 | 972.28 |
| 4 | PMP22 | 0.0103218 | peripheral myelin protein 22 | GTEx | DepMap | Descartes | 8.99 | 1573.97 |
| 5 | CLU | 0.0095567 | clusterin | GTEx | DepMap | Descartes | 14.47 | 1772.86 |
| 6 | TH | 0.0087457 | tyrosine hydroxylase | GTEx | DepMap | Descartes | 5.54 | 940.02 |
| 7 | CNTNAP2 | 0.0085712 | contactin associated protein 2 | GTEx | DepMap | Descartes | 3.68 | 119.16 |
| 8 | DBH | 0.0085132 | dopamine beta-hydroxylase | GTEx | DepMap | Descartes | 6.71 | 778.96 |
| 9 | ELAVL4 | 0.0084525 | ELAV like RNA binding protein 4 | GTEx | DepMap | Descartes | 11.34 | 845.99 |
| 10 | VSTM2A | 0.0081186 | V-set and transmembrane domain containing 2A | GTEx | DepMap | Descartes | 3.30 | 321.13 |
| 11 | RAMP1 | 0.0080911 | receptor activity modifying protein 1 | GTEx | DepMap | Descartes | 9.83 | 3589.17 |
| 12 | CALM2 | 0.0079111 | calmodulin 2 | GTEx | DepMap | Descartes | 56.81 | 3812.05 |
| 13 | TUBA1A | 0.0078693 | tubulin alpha 1a | GTEx | DepMap | Descartes | 93.48 | 14182.12 |
| 14 | MLLT11 | 0.0077011 | MLLT11 transcription factor 7 cofactor | GTEx | DepMap | Descartes | 27.14 | 3321.19 |
| 15 | PHOX2A | 0.0074979 | paired like homeobox 2A | GTEx | DepMap | Descartes | 5.18 | 888.99 |
| 16 | CHGA | 0.0072050 | chromogranin A | GTEx | DepMap | Descartes | 4.25 | 662.30 |
| 17 | GNAS | 0.0071699 | GNAS complex locus | GTEx | DepMap | Descartes | 25.92 | 2033.26 |
| 18 | TMOD1 | 0.0070401 | tropomodulin 1 | GTEx | DepMap | Descartes | 5.07 | 491.72 |
| 19 | UCHL1 | 0.0070231 | ubiquitin C-terminal hydrolase L1 | GTEx | DepMap | Descartes | 12.20 | 2291.55 |
| 20 | FAIM2 | 0.0069029 | Fas apoptotic inhibitory molecule 2 | GTEx | DepMap | Descartes | 2.86 | 195.70 |
| 21 | C1QL1 | 0.0068111 | complement C1q like 1 | GTEx | DepMap | Descartes | 1.56 | 283.35 |
| 22 | STMN2 | 0.0067652 | stathmin 2 | GTEx | DepMap | Descartes | 70.70 | 10962.20 |
| 23 | PCSK1N | 0.0067364 | proprotein convertase subtilisin/kexin type 1 inhibitor | GTEx | DepMap | Descartes | 14.49 | 4392.91 |
| 24 | NREP | 0.0067356 | neuronal regeneration related protein | GTEx | DepMap | Descartes | 11.31 | NA |
| 25 | TUSC3 | 0.0066717 | tumor suppressor candidate 3 | GTEx | DepMap | Descartes | 4.07 | 331.62 |
| 26 | CPNE2 | 0.0065456 | copine 2 | GTEx | DepMap | Descartes | 2.22 | 194.73 |
| 27 | YWHAQ | 0.0065259 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta | GTEx | DepMap | Descartes | 15.11 | 2044.97 |
| 28 | YWHAH | 0.0065059 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein eta | GTEx | DepMap | Descartes | 9.35 | 1463.81 |
| 29 | TUBB2B | 0.0064973 | tubulin beta 2B class IIb | GTEx | DepMap | Descartes | 28.88 | 4404.20 |
| 30 | GFRA3 | 0.0062613 | GDNF family receptor alpha 3 | GTEx | DepMap | Descartes | 2.98 | 458.95 |
| 31 | DNER | 0.0062353 | delta/notch like EGF repeat containing | GTEx | DepMap | Descartes | 1.34 | 137.58 |
| 32 | DDC | 0.0062103 | dopa decarboxylase | GTEx | DepMap | Descartes | 2.60 | 384.59 |
| 33 | SCG5 | 0.0061786 | secretogranin V | GTEx | DepMap | Descartes | 3.29 | 853.39 |
| 34 | CAMK2B | 0.0060742 | calcium/calmodulin dependent protein kinase II beta | GTEx | DepMap | Descartes | 3.25 | 223.13 |
| 35 | GAP43 | 0.0060681 | growth associated protein 43 | GTEx | DepMap | Descartes | 16.43 | 2557.78 |
| 36 | YWHAE | 0.0060452 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon | GTEx | DepMap | Descartes | 13.89 | 1870.62 |
| 37 | TUBB2A | 0.0060213 | tubulin beta 2A class IIa | GTEx | DepMap | Descartes | 19.74 | 3476.18 |
| 38 | MAP1LC3A | 0.0059473 | microtubule associated protein 1 light chain 3 alpha | GTEx | DepMap | Descartes | 5.53 | 1767.56 |
| 39 | MARCKSL1 | 0.0059159 | MARCKS like 1 | GTEx | DepMap | Descartes | 20.38 | 4051.40 |
| 40 | MIF | 0.0058999 | macrophage migration inhibitory factor | GTEx | DepMap | Descartes | 21.91 | 6694.23 |
| 41 | MAP1B | 0.0058948 | microtubule associated protein 1B | GTEx | DepMap | Descartes | 24.19 | 598.30 |
| 42 | QDPR | 0.0058466 | quinoid dihydropteridine reductase | GTEx | DepMap | Descartes | 3.34 | 657.20 |
| 43 | ATP6V1G2 | 0.0058053 | ATPase H+ transporting V1 subunit G2 | GTEx | DepMap | Descartes | 2.51 | 499.22 |
| 44 | GNG3 | 0.0057544 | G protein subunit gamma 3 | GTEx | DepMap | Descartes | 4.39 | 1563.11 |
| 45 | NRCAM | 0.0057528 | neuronal cell adhesion molecule | GTEx | DepMap | Descartes | 1.36 | 71.47 |
| 46 | APLP1 | 0.0057428 | amyloid beta precursor like protein 1 | GTEx | DepMap | Descartes | 3.13 | 420.93 |
| 47 | ARHGAP36 | 0.0056949 | Rho GTPase activating protein 36 | GTEx | DepMap | Descartes | 2.24 | 242.37 |
| 48 | SLC18A1 | 0.0056771 | solute carrier family 18 member A1 | GTEx | DepMap | Descartes | 1.28 | 137.40 |
| 49 | SNAP25 | 0.0056728 | synaptosome associated protein 25 | GTEx | DepMap | Descartes | 3.93 | 525.20 |
| 50 | MAP7 | 0.0056698 | microtubule associated protein 7 | GTEx | DepMap | Descartes | 2.57 | 187.79 |
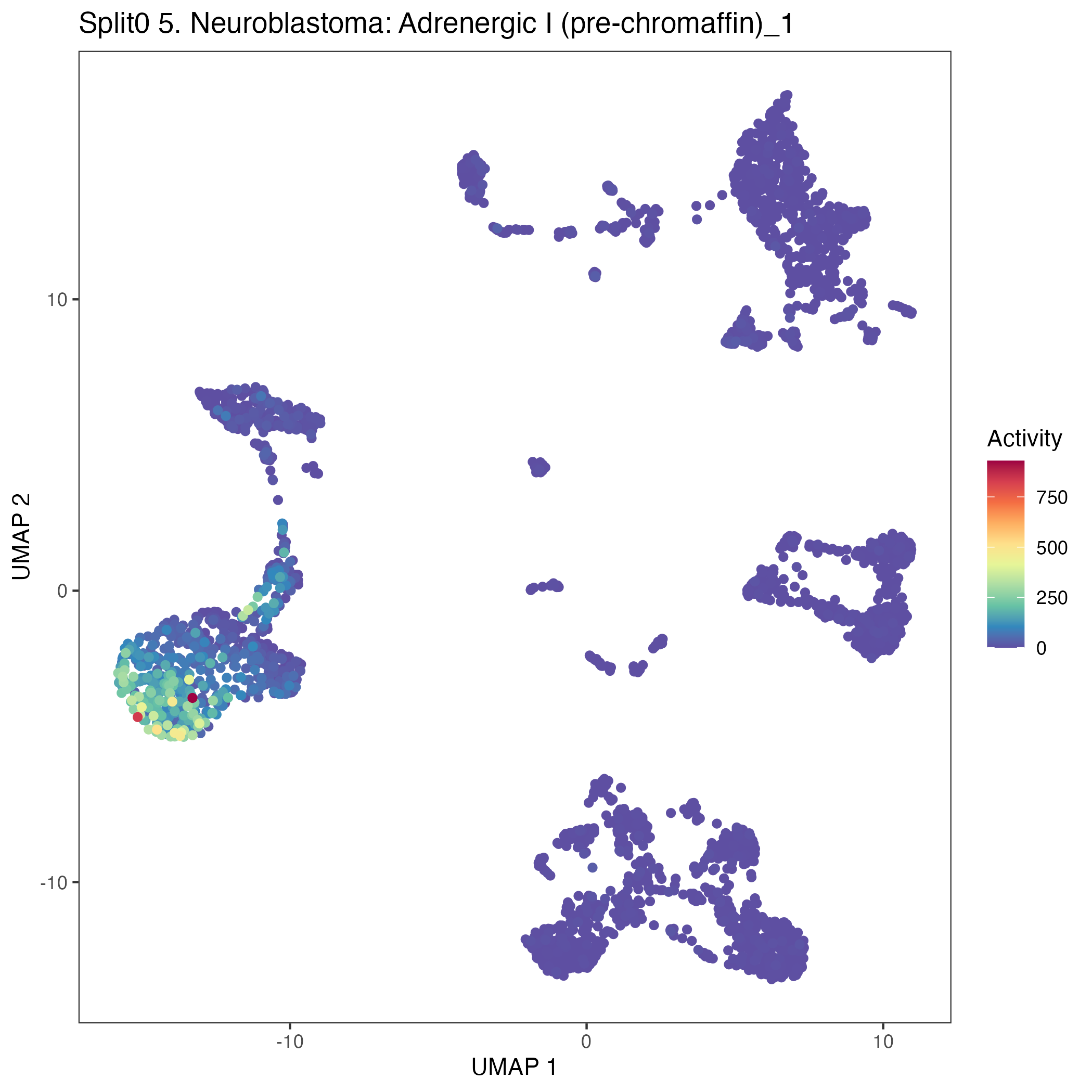
UMAP plots showing activity of gene expression program identified in GEP 5. Neuroblastoma: Adrenergic I (pre-chromaffin):
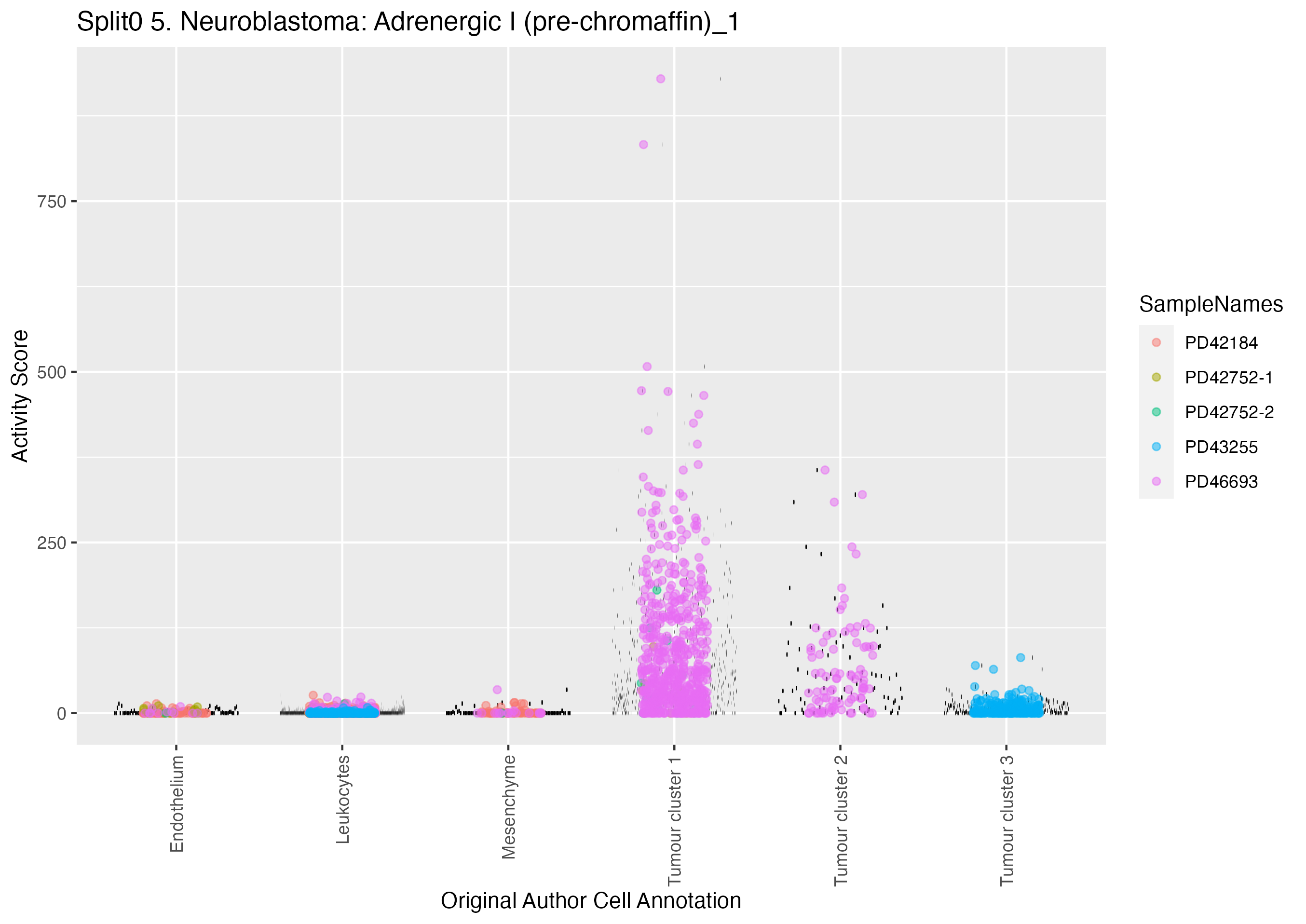
Boxlot showing activity of gene expression program identified in GEP 5. Neuroblastoma: Adrenergic I (pre-chromaffin):
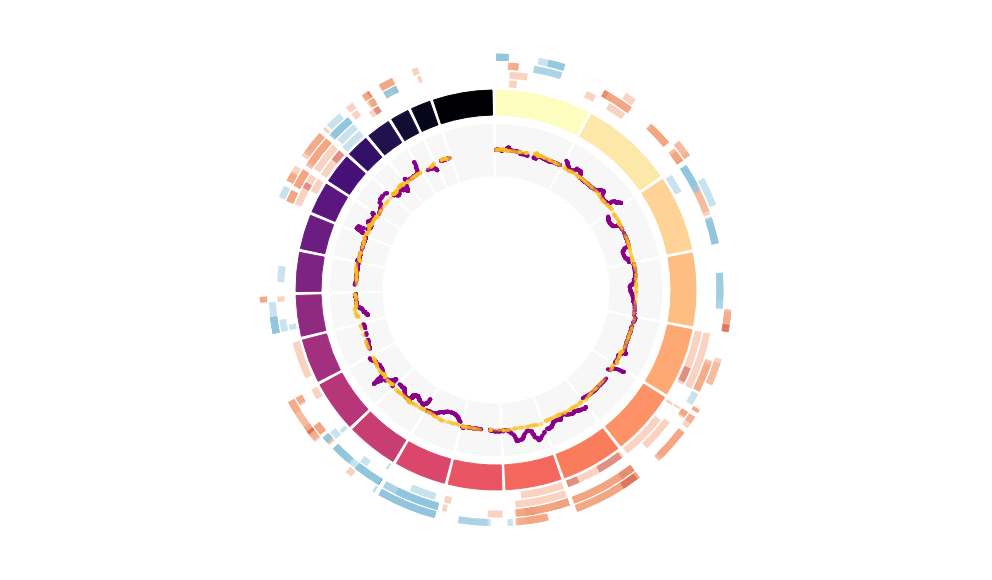
CNV Data procured from inferCNV.
Outer tracks are putative CNV regions (gains = red, losses = blue) for each patient
Inner track is expression data representing:
The top cells expressing this GEP (purple)
Random cells (n =50) from the reference set used in inferCNV (orange)
Gene set Enrichments for this program, caculated from top 50 genes
mSigDB Cell Types Gene Set:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| DURANTE_ADULT_OLFACTORY_NEUROEPITHELIUM_GLOBOSE_BASAL_CELLS | 6.24e-11 | 72.47 | 25.33 | 2.79e-09 | 4.19e-08 | 7ELAVL4, TUBA1A, UCHL1, STMN2, TUBB2B, MARCKSL1, MAP1B |
35 |
| DESCARTES_FETAL_INTESTINE_ENS_NEURONS | 2.50e-17 | 36.92 | 18.32 | 8.39e-15 | 1.68e-14 | 15TH, CNTNAP2, DBH, ELAVL4, MLLT11, PHOX2A, UCHL1, STMN2, TUBB2B, DNER, GAP43, TUBB2A, MAP1B, ATP6V1G2, GNG3 |
160 |
| TRAVAGLINI_LUNG_NEUROENDOCRINE_CELL | 1.58e-15 | 38.15 | 18.12 | 1.32e-13 | 1.06e-12 | 13PKIB, ELAVL4, UCHL1, STMN2, PCSK1N, TUBB2B, GFRA3, DDC, SCG5, CAMK2B, MAP1B, APLP1, SNAP25 |
128 |
| DURANTE_ADULT_OLFACTORY_NEUROEPITHELIUM_IMMATURE_NEURONS | 4.65e-15 | 34.83 | 16.57 | 3.47e-13 | 3.12e-12 | 13ELAVL4, CALM2, TUBA1A, UCHL1, STMN2, PCSK1N, TUBB2B, SCG5, GAP43, TUBB2A, MARCKSL1, MAP1B, APLP1 |
139 |
| HU_FETAL_RETINA_HORIZONTAL | 9.97e-09 | 49.98 | 16.33 | 3.52e-07 | 6.69e-06 | 6NTRK1, CNTNAP2, TUBA1A, MLLT11, STMN2, TUBB2B |
40 |
| DESCARTES_FETAL_PANCREAS_ENS_NEURONS | 1.16e-15 | 32.61 | 15.90 | 1.11e-13 | 7.80e-13 | 14CNTNAP2, DBH, ELAVL4, TUBA1A, MLLT11, PHOX2A, UCHL1, STMN2, TUBB2B, DNER, GAP43, TUBB2A, MAP1B, GNG3 |
163 |
| BUSSLINGER_GASTRIC_ANTRAL_ECS | 1.27e-05 | 88.17 | 14.90 | 2.37e-04 | 8.54e-03 | 3PCSK1N, DDC, SCG5 |
12 |
| BUSSLINGER_GASTRIC_OXYNTIC_ENTEROCHROMAFFIN_LIKE_CELLS | 3.66e-11 | 36.05 | 14.88 | 1.76e-09 | 2.46e-08 | 9CHGA, PCSK1N, YWHAQ, GFRA3, DNER, SCG5, CAMK2B, QDPR, APLP1 |
85 |
| MANNO_MIDBRAIN_NEUROTYPES_HOMTN | 1.95e-19 | 24.61 | 13.21 | 1.31e-16 | 1.31e-16 | 21NTRK1, CNTNAP2, DBH, ELAVL4, VSTM2A, TUBA1A, MLLT11, PHOX2A, CHGA, UCHL1, FAIM2, STMN2, TUBB2B, SCG5, GAP43, TUBB2A, MAP1LC3A, MAP1B, GNG3, APLP1, SNAP25 |
389 |
| DURANTE_ADULT_OLFACTORY_NEUROEPITHELIUM_MATURE_NEURONS | 2.14e-13 | 25.23 | 12.09 | 1.31e-11 | 1.44e-10 | 13CALM2, TUBA1A, CHGA, UCHL1, FAIM2, STMN2, PCSK1N, TUBB2B, DNER, GAP43, MAP1B, APLP1, SNAP25 |
187 |
| DESCARTES_MAIN_FETAL_SYMPATHOBLASTS | 8.89e-08 | 33.37 | 11.13 | 2.71e-06 | 5.97e-05 | 6TH, DBH, CHGA, C1QL1, QDPR, SLC18A1 |
57 |
| DESCARTES_FETAL_STOMACH_ENS_NEURONS | 1.45e-08 | 30.33 | 11.11 | 4.65e-07 | 9.76e-06 | 7CNTNAP2, ELAVL4, UCHL1, STMN2, TUBB2B, GAP43, TUBB2A |
74 |
| BUSSLINGER_GASTRIC_D_CELLS | 4.22e-06 | 45.22 | 10.96 | 8.84e-05 | 2.83e-03 | 4PCSK1N, SCG5, CAMK2B, MAP1B |
28 |
| HU_FETAL_RETINA_RGC | 5.42e-17 | 19.71 | 10.52 | 9.09e-15 | 3.64e-14 | 20NTRK1, ELAVL4, CALM2, TUBA1A, MLLT11, UCHL1, STMN2, PCSK1N, YWHAQ, YWHAH, TUBB2B, DNER, SCG5, GAP43, TUBB2A, MAP1B, ATP6V1G2, NRCAM, APLP1, SNAP25 |
443 |
| MANNO_MIDBRAIN_NEUROTYPES_HDA2 | 4.67e-17 | 18.41 | 9.91 | 9.09e-15 | 3.13e-14 | 21TH, CNTNAP2, ELAVL4, VSTM2A, TUBA1A, MLLT11, UCHL1, FAIM2, C1QL1, STMN2, NREP, TUBB2B, DDC, SCG5, GAP43, TUBB2A, MAP1B, GNG3, APLP1, SLC18A1, SNAP25 |
513 |
| HU_FETAL_RETINA_AMACRINE | 1.79e-07 | 29.35 | 9.84 | 4.81e-06 | 1.20e-04 | 6MLLT11, STMN2, NREP, TUBB2B, TUBB2A, MARCKSL1 |
64 |
| BUSSLINGER_DUODENAL_EC_CELLS | 6.42e-06 | 40.15 | 9.82 | 1.27e-04 | 4.31e-03 | 4CHGA, PCSK1N, DDC, MAP1B |
31 |
| MANNO_MIDBRAIN_NEUROTYPES_HDA | 6.47e-16 | 17.16 | 9.17 | 7.24e-14 | 4.34e-13 | 20TH, ELAVL4, TUBA1A, MLLT11, CHGA, UCHL1, C1QL1, STMN2, NREP, TUBB2B, DNER, DDC, SCG5, CAMK2B, GAP43, MAP1B, GNG3, APLP1, SLC18A1, SNAP25 |
506 |
| MANNO_MIDBRAIN_NEUROTYPES_HDA1 | 5.74e-16 | 16.10 | 8.66 | 7.24e-14 | 3.85e-13 | 21TH, ELAVL4, VSTM2A, MLLT11, CHGA, UCHL1, FAIM2, STMN2, PCSK1N, NREP, TUBB2B, DNER, DDC, SCG5, CAMK2B, GAP43, MAP1B, GNG3, APLP1, SLC18A1, SNAP25 |
584 |
| MANNO_MIDBRAIN_NEUROTYPES_HSERT | 2.37e-14 | 16.29 | 8.54 | 1.59e-12 | 1.59e-11 | 18VSTM2A, MLLT11, CHGA, UCHL1, STMN2, PCSK1N, YWHAH, DDC, SCG5, CAMK2B, GAP43, TUBB2A, MAP1LC3A, MAP1B, QDPR, GNG3, APLP1, SNAP25 |
450 |
Dowload full table
mSigDB Hallmark Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| HALLMARK_KRAS_SIGNALING_UP | 7.71e-03 | 5.55 | 1.44 | 3.85e-01 | 3.85e-01 | 4PCSK1N, SCG5, SNAP25, MAP7 |
200 |
| HALLMARK_SPERMATOGENESIS | 1.61e-02 | 6.04 | 1.19 | 4.01e-01 | 8.03e-01 | 3PCSK1N, SCG5, MAP7 |
135 |
| HALLMARK_UV_RESPONSE_DN | 1.10e-01 | 3.67 | 0.43 | 9.24e-01 | 1.00e+00 | 2PMP22, MAP1B |
144 |
| HALLMARK_FATTY_ACID_METABOLISM | 1.28e-01 | 3.34 | 0.39 | 9.24e-01 | 1.00e+00 | 2YWHAH, MIF |
158 |
| HALLMARK_MYOGENESIS | 1.85e-01 | 2.63 | 0.31 | 9.24e-01 | 1.00e+00 | 2CLU, CAMK2B |
200 |
| HALLMARK_MTORC1_SIGNALING | 1.85e-01 | 2.63 | 0.31 | 9.24e-01 | 1.00e+00 | 2MLLT11, QDPR |
200 |
| HALLMARK_MYC_TARGETS_V1 | 1.85e-01 | 2.63 | 0.31 | 9.24e-01 | 1.00e+00 | 2YWHAQ, YWHAE |
200 |
| HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | 1.85e-01 | 2.63 | 0.31 | 9.24e-01 | 1.00e+00 | 2PMP22, APLP1 |
200 |
| HALLMARK_HEDGEHOG_SIGNALING | 1.34e-01 | 7.29 | 0.18 | 9.24e-01 | 1.00e+00 | 1NRCAM |
36 |
| HALLMARK_PANCREAS_BETA_CELLS | 1.47e-01 | 6.54 | 0.16 | 9.24e-01 | 1.00e+00 | 1CHGA |
40 |
| HALLMARK_CHOLESTEROL_HOMEOSTASIS | 2.55e-01 | 3.49 | 0.09 | 9.77e-01 | 1.00e+00 | 1CLU |
74 |
| HALLMARK_PROTEIN_SECRETION | 3.17e-01 | 2.68 | 0.07 | 9.77e-01 | 1.00e+00 | 1GNAS |
96 |
| HALLMARK_ANDROGEN_RESPONSE | 3.28e-01 | 2.58 | 0.06 | 9.77e-01 | 1.00e+00 | 1MAP7 |
100 |
| HALLMARK_PEROXISOME | 3.39e-01 | 2.48 | 0.06 | 9.77e-01 | 1.00e+00 | 1YWHAH |
104 |
| HALLMARK_UNFOLDED_PROTEIN_RESPONSE | 3.62e-01 | 2.28 | 0.06 | 9.77e-01 | 1.00e+00 | 1TUBB2A |
113 |
| HALLMARK_COAGULATION | 4.22e-01 | 1.86 | 0.05 | 9.77e-01 | 1.00e+00 | 1CLU |
138 |
| HALLMARK_APOPTOSIS | 4.72e-01 | 1.59 | 0.04 | 9.77e-01 | 1.00e+00 | 1CLU |
161 |
| HALLMARK_MITOTIC_SPINDLE | 5.45e-01 | 1.29 | 0.03 | 9.77e-01 | 1.00e+00 | 1YWHAE |
199 |
| HALLMARK_IL2_STAT5_SIGNALING | 5.45e-01 | 1.29 | 0.03 | 9.77e-01 | 1.00e+00 | 1APLP1 |
199 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 5.47e-01 | 1.28 | 0.03 | 9.77e-01 | 1.00e+00 | 1TUBB2A |
200 |
Dowload full table
KEGG Pathways:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| KEGG_TYROSINE_METABOLISM | 2.20e-05 | 28.56 | 7.12 | 2.05e-03 | 4.10e-03 | 4TH, DBH, DDC, MIF |
42 |
| KEGG_PATHOGENIC_ESCHERICHIA_COLI_INFECTION | 6.91e-05 | 20.88 | 5.27 | 3.70e-03 | 1.29e-02 | 4TUBA1A, YWHAQ, TUBB2B, TUBB2A |
56 |
| KEGG_NEUROTROPHIN_SIGNALING_PATHWAY | 9.51e-06 | 14.20 | 4.85 | 1.77e-03 | 1.77e-03 | 6NTRK1, CALM2, YWHAQ, YWHAH, CAMK2B, YWHAE |
126 |
| KEGG_OOCYTE_MEIOSIS | 7.97e-05 | 12.85 | 3.91 | 3.70e-03 | 1.48e-02 | 5CALM2, YWHAQ, YWHAH, CAMK2B, YWHAE |
113 |
| KEGG_PHENYLALANINE_METABOLISM | 2.28e-03 | 32.49 | 3.53 | 7.06e-02 | 4.24e-01 | 2DDC, MIF |
18 |
| KEGG_GAP_JUNCTION | 4.31e-04 | 12.63 | 3.23 | 1.60e-02 | 8.02e-02 | 4TUBA1A, GNAS, TUBB2B, TUBB2A |
90 |
| KEGG_GNRH_SIGNALING_PATHWAY | 7.38e-03 | 8.14 | 1.59 | 1.72e-01 | 1.00e+00 | 3CALM2, GNAS, CAMK2B |
101 |
| KEGG_MELANOGENESIS | 7.38e-03 | 8.14 | 1.59 | 1.72e-01 | 1.00e+00 | 3CALM2, GNAS, CAMK2B |
101 |
| KEGG_VASCULAR_SMOOTH_MUSCLE_CONTRACTION | 1.05e-02 | 7.12 | 1.40 | 2.17e-01 | 1.00e+00 | 3RAMP1, CALM2, GNAS |
115 |
| KEGG_CELL_CYCLE | 1.31e-02 | 6.54 | 1.28 | 2.44e-01 | 1.00e+00 | 3YWHAQ, YWHAH, YWHAE |
125 |
| KEGG_PARKINSONS_DISEASE | 1.45e-02 | 6.28 | 1.23 | 2.46e-01 | 1.00e+00 | 3TH, UCHL1, SLC18A1 |
130 |
| KEGG_TASTE_TRANSDUCTION | 1.80e-02 | 10.41 | 1.19 | 2.77e-01 | 1.00e+00 | 2GNAS, GNG3 |
52 |
| KEGG_VIBRIO_CHOLERAE_INFECTION | 1.94e-02 | 10.01 | 1.15 | 2.77e-01 | 1.00e+00 | 2GNAS, ATP6V1G2 |
54 |
| KEGG_GLIOMA | 2.73e-02 | 8.26 | 0.95 | 3.63e-01 | 1.00e+00 | 2CALM2, CAMK2B |
65 |
| KEGG_CALCIUM_SIGNALING_PATHWAY | 3.26e-02 | 4.56 | 0.90 | 3.79e-01 | 1.00e+00 | 3CALM2, GNAS, CAMK2B |
178 |
| KEGG_LONG_TERM_POTENTIATION | 3.13e-02 | 7.66 | 0.88 | 3.79e-01 | 1.00e+00 | 2CALM2, CAMK2B |
70 |
| KEGG_FOLATE_BIOSYNTHESIS | 4.29e-02 | 25.48 | 0.58 | 4.70e-01 | 1.00e+00 | 1QDPR |
11 |
| KEGG_CELL_ADHESION_MOLECULES_CAMS | 9.66e-02 | 3.98 | 0.46 | 9.98e-01 | 1.00e+00 | 2CNTNAP2, NRCAM |
133 |
| KEGG_HISTIDINE_METABOLISM | 1.09e-01 | 9.11 | 0.22 | 1.00e+00 | 1.00e+00 | 1DDC |
29 |
| KEGG_THYROID_CANCER | 1.09e-01 | 9.11 | 0.22 | 1.00e+00 | 1.00e+00 | 1NTRK1 |
29 |
Dowload full table
CHR Positional Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| chr6p25 | 4.17e-02 | 6.51 | 0.75 | 1.00e+00 | 1.00e+00 | 2TUBB2B, TUBB2A |
82 |
| chr8p21 | 9.06e-02 | 4.13 | 0.48 | 1.00e+00 | 1.00e+00 | 2CLU, SLC18A1 |
128 |
| chr1q23 | 2.09e-01 | 2.42 | 0.28 | 1.00e+00 | 1.00e+00 | 2RGS5, NTRK1 |
217 |
| chr4p13 | 1.16e-01 | 8.50 | 0.20 | 1.00e+00 | 1.00e+00 | 1UCHL1 |
31 |
| chr16q13 | 1.47e-01 | 6.54 | 0.16 | 1.00e+00 | 1.00e+00 | 1CPNE2 |
40 |
| chr12q13 | 6.71e-01 | 1.29 | 0.15 | 1.00e+00 | 1.00e+00 | 2TUBA1A, FAIM2 |
407 |
| chr8p22 | 1.64e-01 | 5.80 | 0.14 | 1.00e+00 | 1.00e+00 | 1TUSC3 |
45 |
| chr7p13 | 1.81e-01 | 5.21 | 0.13 | 1.00e+00 | 1.00e+00 | 1CAMK2B |
50 |
| chr5q22 | 1.97e-01 | 4.72 | 0.12 | 1.00e+00 | 1.00e+00 | 1NREP |
55 |
| chr7p12 | 2.06e-01 | 4.47 | 0.11 | 1.00e+00 | 1.00e+00 | 1DDC |
58 |
| chr1p33 | 2.13e-01 | 4.32 | 0.11 | 1.00e+00 | 1.00e+00 | 1ELAVL4 |
60 |
| chr17p12 | 2.22e-01 | 4.11 | 0.10 | 1.00e+00 | 1.00e+00 | 1PMP22 |
63 |
| chr7q35 | 2.28e-01 | 3.99 | 0.10 | 1.00e+00 | 1.00e+00 | 1CNTNAP2 |
65 |
| chr2p21 | 2.73e-01 | 3.23 | 0.08 | 1.00e+00 | 1.00e+00 | 1CALM2 |
80 |
| chr2q36 | 2.78e-01 | 3.15 | 0.08 | 1.00e+00 | 1.00e+00 | 1DNER |
82 |
| chr7p11 | 2.78e-01 | 3.15 | 0.08 | 1.00e+00 | 1.00e+00 | 1VSTM2A |
82 |
| chr15q13 | 2.93e-01 | 2.97 | 0.07 | 1.00e+00 | 1.00e+00 | 1SCG5 |
87 |
| chr20p12 | 3.39e-01 | 2.48 | 0.06 | 1.00e+00 | 1.00e+00 | 1SNAP25 |
104 |
| chr6q23 | 3.44e-01 | 2.43 | 0.06 | 1.00e+00 | 1.00e+00 | 1MAP7 |
106 |
| chr2p25 | 3.72e-01 | 2.20 | 0.05 | 1.00e+00 | 1.00e+00 | 1YWHAQ |
117 |
Dowload full table
Transcription Factor Targets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GATA_Q6 | 1.23e-05 | 10.49 | 3.93 | 1.39e-02 | 1.39e-02 | 7CLU, ELAVL4, PHOX2A, NREP, TUBB2B, GFRA3, SLC18A1 |
201 |
| NRSF_01 | 5.72e-04 | 11.68 | 2.99 | 9.25e-02 | 6.48e-01 | 4CHGA, STMN2, DNER, SNAP25 |
97 |
| E4BP4_01 | 2.22e-04 | 7.85 | 2.71 | 6.28e-02 | 2.51e-01 | 6ELAVL4, CALM2, TUBA1A, UCHL1, FAIM2, NREP |
223 |
| MYB_Q3 | 4.31e-04 | 6.90 | 2.38 | 8.64e-02 | 4.88e-01 | 6CHGA, GNAS, PCSK1N, YWHAQ, YWHAE, GNG3 |
253 |
| PAX4_03 | 4.58e-04 | 6.82 | 2.35 | 8.64e-02 | 5.19e-01 | 6C1QL1, STMN2, PCSK1N, NREP, YWHAE, MAP1B |
256 |
| GGGYGTGNY_UNKNOWN | 2.04e-04 | 4.63 | 2.05 | 6.28e-02 | 2.31e-01 | 10PMP22, CALM2, TUBA1A, MLLT11, UCHL1, TUSC3, YWHAH, GAP43, MAP1B, ARHGAP36 |
686 |
| AP2ALPHA_01 | 2.51e-03 | 5.84 | 1.79 | 2.29e-01 | 1.00e+00 | 5MAP1LC3A, MARCKSL1, MAP1B, APLP1, ARHGAP36 |
243 |
| LMO2COM_02 | 2.55e-03 | 5.81 | 1.78 | 2.29e-01 | 1.00e+00 | 5CLU, ELAVL4, VSTM2A, CALM2, GFRA3 |
244 |
| PTF1BETA_Q6 | 2.64e-03 | 5.76 | 1.77 | 2.29e-01 | 1.00e+00 | 5NTRK1, PMP22, YWHAE, MARCKSL1, GNG3 |
246 |
| TTCYRGAA_UNKNOWN | 1.93e-03 | 5.10 | 1.77 | 2.29e-01 | 1.00e+00 | 6CLU, CNTNAP2, MLLT11, UCHL1, YWHAQ, CAMK2B |
340 |
| TGGAAA_NFAT_Q4_01 | 1.59e-04 | 3.36 | 1.75 | 6.28e-02 | 1.80e-01 | 17NTRK1, PMP22, CNTNAP2, ELAVL4, TUBA1A, GNAS, UCHL1, C1QL1, STMN2, NREP, YWHAQ, DDC, GAP43, MARCKSL1, MAP1B, SNAP25, MAP7 |
1934 |
| TGACCTTG_SF1_Q6 | 2.97e-03 | 5.60 | 1.72 | 2.29e-01 | 1.00e+00 | 5CHGA, UCHL1, YWHAH, DNER, MAP1B |
253 |
| E47_01 | 3.07e-03 | 5.56 | 1.71 | 2.29e-01 | 1.00e+00 | 5ELAVL4, MLLT11, GNAS, TUBB2B, ARHGAP36 |
255 |
| CREBP1_Q2 | 3.33e-03 | 5.45 | 1.67 | 2.29e-01 | 1.00e+00 | 5TH, CALM2, GNAS, MAP1LC3A, SNAP25 |
260 |
| PAX2_02 | 3.61e-03 | 5.34 | 1.64 | 2.29e-01 | 1.00e+00 | 5PMP22, ELAVL4, CALM2, STMN2, DNER |
265 |
| DR4_Q2 | 3.67e-03 | 5.32 | 1.64 | 2.29e-01 | 1.00e+00 | 5ELAVL4, PCSK1N, NREP, MAP1LC3A, ARHGAP36 |
266 |
| GTTRYCATRR_UNKNOWN | 4.96e-03 | 6.32 | 1.63 | 2.81e-01 | 1.00e+00 | 4FAIM2, PCSK1N, YWHAQ, YWHAE |
176 |
| CREB_01 | 3.79e-03 | 5.28 | 1.62 | 2.29e-01 | 1.00e+00 | 5TH, CALM2, TUBB2B, MAP1LC3A, SNAP25 |
268 |
| MYB_Q5_01 | 3.84e-03 | 5.26 | 1.62 | 2.29e-01 | 1.00e+00 | 5ELAVL4, GNAS, PCSK1N, YWHAE, GNG3 |
269 |
| CREBP1_01 | 5.68e-03 | 6.07 | 1.57 | 2.94e-01 | 1.00e+00 | 4ELAVL4, CALM2, NREP, TUBB2B |
183 |
Dowload full table
GO Biological Processes:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GOBP_NEUROTRANSMITTER_LOADING_INTO_SYNAPTIC_VESICLE | 3.28e-06 | 158.40 | 23.90 | 6.92e-03 | 2.45e-02 | 3TH, DDC, SLC18A1 |
8 |
| GOBP_RESPONSE_TO_INSECTICIDE | 4.91e-06 | 132.55 | 20.77 | 7.34e-03 | 3.67e-02 | 3TH, DDC, MAP1B |
9 |
| GOBP_ALKALOID_METABOLIC_PROCESS | 3.22e-04 | 103.49 | 9.64 | 1.03e-01 | 1.00e+00 | 2TH, DDC |
7 |
| GOBP_NOREPINEPHRINE_BIOSYNTHETIC_PROCESS | 3.22e-04 | 103.49 | 9.64 | 1.03e-01 | 1.00e+00 | 2TH, DBH |
7 |
| GOBP_CATECHOL_CONTAINING_COMPOUND_BIOSYNTHETIC_PROCESS | 6.44e-05 | 46.77 | 8.51 | 4.38e-02 | 4.82e-01 | 3TH, DBH, DDC |
20 |
| GOBP_SYMPATHETIC_NERVOUS_SYSTEM_DEVELOPMENT | 7.49e-05 | 44.24 | 8.07 | 4.67e-02 | 5.60e-01 | 3NTRK1, PHOX2A, GFRA3 |
21 |
| GOBP_NEURAL_NUCLEUS_DEVELOPMENT | 3.70e-06 | 25.20 | 7.53 | 6.92e-03 | 2.77e-02 | 5CALM2, PHOX2A, YWHAQ, YWHAH, YWHAE |
60 |
| GOBP_CELLULAR_RESPONSE_TO_NICOTINE | 5.49e-04 | 74.10 | 7.33 | 1.37e-01 | 1.00e+00 | 2NTRK1, TH |
9 |
| GOBP_SUBSTANTIA_NIGRA_DEVELOPMENT | 2.42e-05 | 27.83 | 6.94 | 2.26e-02 | 1.81e-01 | 4CALM2, YWHAQ, YWHAH, YWHAE |
43 |
| GOBP_DOPAMINE_BIOSYNTHETIC_PROCESS | 8.35e-04 | 57.69 | 5.91 | 1.64e-01 | 1.00e+00 | 2TH, DDC |
11 |
| GOBP_NEURONAL_ION_CHANNEL_CLUSTERING | 8.35e-04 | 57.69 | 5.91 | 1.64e-01 | 1.00e+00 | 2CNTNAP2, NRCAM |
11 |
| GOBP_PROTEIN_INSERTION_INTO_MITOCHONDRIAL_MEMBRANE_INVOLVED_IN_APOPTOTIC_SIGNALING_PATHWAY | 2.22e-04 | 29.50 | 5.54 | 8.90e-02 | 1.00e+00 | 3YWHAQ, YWHAH, YWHAE |
30 |
| GOBP_NEURON_MIGRATION | 1.45e-06 | 14.74 | 5.50 | 5.44e-03 | 1.09e-02 | 7TUBB2B, GFRA3, DNER, CAMK2B, TUBB2A, MAP1B, NRCAM |
145 |
| GOBP_MIDBRAIN_DEVELOPMENT | 2.16e-05 | 17.14 | 5.17 | 2.26e-02 | 1.62e-01 | 5CALM2, PHOX2A, YWHAQ, YWHAH, YWHAE |
86 |
| GOBP_NOREPINEPHRINE_METABOLIC_PROCESS | 1.18e-03 | 47.19 | 4.96 | 2.12e-01 | 1.00e+00 | 2TH, DBH |
13 |
| GOBP_POSITIVE_REGULATION_OF_MITOCHONDRIAL_OUTER_MEMBRANE_PERMEABILIZATION_INVOLVED_IN_APOPTOTIC_SIGNALING_PATHWAY | 3.53e-04 | 24.91 | 4.72 | 1.06e-01 | 1.00e+00 | 3YWHAQ, YWHAH, YWHAE |
35 |
| GOBP_RESPONSE_TO_WATER | 1.37e-03 | 43.30 | 4.59 | 2.28e-01 | 1.00e+00 | 2NTRK1, TH |
14 |
| GOBP_DOPAMINE_METABOLIC_PROCESS | 4.51e-04 | 22.75 | 4.33 | 1.25e-01 | 1.00e+00 | 3TH, DBH, DDC |
38 |
| GOBP_TOXIN_METABOLIC_PROCESS | 1.58e-03 | 39.95 | 4.27 | 2.36e-01 | 1.00e+00 | 2TH, DDC |
15 |
| GOBP_AMINE_BIOSYNTHETIC_PROCESS | 4.87e-04 | 22.12 | 4.22 | 1.30e-01 | 1.00e+00 | 3TH, DBH, DDC |
39 |
Dowload full table
Immunological Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GSE17721_CTRL_VS_GARDIQUIMOD_0.5H_BMDC_DN | 1.20e-04 | 8.83 | 3.04 | 3.01e-01 | 5.86e-01 | 6TUBA1A, GNAS, C1QL1, CAMK2B, MARCKSL1, SLC18A1 |
199 |
| GSE2405_HEAT_KILLED_LYSATE_VS_LIVE_A_PHAGOCYTOPHILUM_STIM_NEUTROPHIL_24H_UP | 1.24e-04 | 8.78 | 3.02 | 3.01e-01 | 6.03e-01 | 6RGS5, PMP22, CLU, SCG5, MAP1LC3A, MARCKSL1 |
200 |
| GSE21546_UNSTIM_VS_ANTI_CD3_STIM_SAP1A_KO_DP_THYMOCYTES_UP | 1.03e-03 | 7.19 | 2.20 | 1.00e+00 | 1.00e+00 | 5MLLT11, UCHL1, NREP, SCG5, TUBB2A |
198 |
| GSE22025_UNTREATED_VS_PROGESTERONE_TREATED_CD4_TCELL_DN | 1.06e-03 | 7.16 | 2.19 | 1.00e+00 | 1.00e+00 | 5CALM2, STMN2, TUSC3, YWHAQ, YWHAH |
199 |
| GSE360_T_GONDII_VS_B_MALAYI_LOW_DOSE_DC_UP | 7.32e-03 | 5.63 | 1.46 | 1.00e+00 | 1.00e+00 | 4PMP22, FAIM2, TUSC3, TUBB2A |
197 |
| GSE24634_IL4_VS_CTRL_TREATED_NAIVE_CD4_TCELL_DAY10_UP | 7.45e-03 | 5.60 | 1.45 | 1.00e+00 | 1.00e+00 | 4RAMP1, MLLT11, MARCKSL1, SNAP25 |
198 |
| GSE360_CTRL_VS_B_MALAYI_HIGH_DOSE_DC_DN | 7.45e-03 | 5.60 | 1.45 | 1.00e+00 | 1.00e+00 | 4NTRK1, STMN2, TUBB2A, QDPR |
198 |
| GSE360_DC_VS_MAC_T_GONDII_UP | 7.45e-03 | 5.60 | 1.45 | 1.00e+00 | 1.00e+00 | 4PMP22, RAMP1, TUBA1A, SNAP25 |
198 |
| GSE360_T_GONDII_VS_B_MALAYI_HIGH_DOSE_DC_UP | 7.45e-03 | 5.60 | 1.45 | 1.00e+00 | 1.00e+00 | 4PMP22, TUSC3, CAMK2B, SNAP25 |
198 |
| GSE3982_MAC_VS_TH1_UP | 7.45e-03 | 5.60 | 1.45 | 1.00e+00 | 1.00e+00 | 4PMP22, TUBA1A, SCG5, TUBB2A |
198 |
| GSE1460_CD4_THYMOCYTE_VS_NAIVE_CD4_TCELL_ADULT_BLOOD_UP | 7.58e-03 | 5.57 | 1.44 | 1.00e+00 | 1.00e+00 | 4MLLT11, TUSC3, YWHAQ, MARCKSL1 |
199 |
| GSE20366_EX_VIVO_VS_DEC205_CONVERSION_NAIVE_CD4_TCELL_DN | 7.58e-03 | 5.57 | 1.44 | 1.00e+00 | 1.00e+00 | 4TUBA1A, SCG5, MAP1LC3A, MARCKSL1 |
199 |
| GSE3982_MAST_CELL_VS_DC_UP | 7.58e-03 | 5.57 | 1.44 | 1.00e+00 | 1.00e+00 | 4NTRK1, PMP22, TMOD1, NREP |
199 |
| GSE7460_CTRL_VS_TGFB_TREATED_ACT_TCONV_UP | 7.58e-03 | 5.57 | 1.44 | 1.00e+00 | 1.00e+00 | 4RGS5, TUBA1A, DDC, TUBB2A |
199 |
| GSE4142_PLASMA_CELL_VS_MEMORY_BCELL_UP | 7.58e-03 | 5.57 | 1.44 | 1.00e+00 | 1.00e+00 | 4RGS5, PHOX2A, MAP1B, SLC18A1 |
199 |
| GSE6674_CPG_VS_CPG_AND_ANTI_IGM_STIM_BCELL_UP | 7.58e-03 | 5.57 | 1.44 | 1.00e+00 | 1.00e+00 | 4TUBA1A, TUBB2B, TUBB2A, MARCKSL1 |
199 |
| GSE28726_NAIVE_CD4_TCELL_VS_NAIVE_NKTCELL_UP | 7.58e-03 | 5.57 | 1.44 | 1.00e+00 | 1.00e+00 | 4CALM2, TUBA1A, YWHAE, TUBB2A |
199 |
| GSE43863_DAY6_EFF_VS_DAY150_MEM_TFH_CD4_TCELL_UP | 7.58e-03 | 5.57 | 1.44 | 1.00e+00 | 1.00e+00 | 4PMP22, CHGA, TUBB2B, MARCKSL1 |
199 |
| GSE13306_TREG_RA_VS_TCONV_RA_UP | 7.71e-03 | 5.55 | 1.44 | 1.00e+00 | 1.00e+00 | 4NTRK1, CNTNAP2, CPNE2, MAP1B |
200 |
| GSE13306_RA_VS_UNTREATED_MEM_CD4_TCELL_DN | 7.71e-03 | 5.55 | 1.44 | 1.00e+00 | 1.00e+00 | 4PKIB, NREP, CAMK2B, MARCKSL1 |
200 |
Top Ranked Transcription Factors for this Gene Expression Program:
| Gene Symbol | TF Rank | DNA Binding Domain | Motif Status | IUPAC PWM | GTEx | DepMap | Decartes |
|---|---|---|---|---|---|---|---|
| NTRK1 | 2 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Membrane bound receptor protein |
| CLU | 5 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Included only because GO says positive regulation of NF-kappaB transcription factor activity, but there is no DBD and no evidence of DNA binding. |
| MLLT11 | 14 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| PHOX2A | 15 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| GATA2 | 73 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| HAND1 | 89 | Yes | Known motif | Obligate heteromer | In vivo/Misc source | Only known motifs are from Transfac or HocoMoco - origin is uncertain | Obligate heteromer (PMID: 10611232). |
| TFAP2B | 94 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| RORB | 101 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| THRA | 103 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | Likely binds as a heterodimer too |
| PCBD1 | 155 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Coactivator for HNF1A-dependent transcription. Regulates the dimerization of homeodomain protein HNF1A and enhances its transcriptional activity (UniProt: P61457) |
| SOX4 | 156 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | Also binds ssDNA loops. |
| HDAC2 | 166 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | Only known motifs are from Transfac or HocoMoco - origin is uncertain | Histone deacetylase; likely to be a transcriptional cofactor |
| TERF2IP | 178 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Contains a single Myb-like domain. Forms a complex with TERF2, where it modulates the binding of TERF2 to telomeres (PMID: 25675958). Unclear if TERF2IP contacts DNA directly. |
| NCOA7 | 184 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Known co-activator; included only because TF-cat documents this |
| HSBP1 | 197 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Protein that has been shown to bind to HSF1 leading to inhibition of its trimerization and DNA-binding( PMID: 9649501). |
| GATA3 | 199 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| HOXC9 | 207 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| RGS9 | 218 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| HAND2 | 241 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| PCBP3 | 261 | No | ssDNA/RNA binding | Not a DNA binding protein | No motif | None | KH domain suggests that this is an RNA-binding protein |
QQ Plot showing correlations with other GEPs in this dataset, calculated by Spearman correlation:
Interactive QQ-plot of gene loadings:
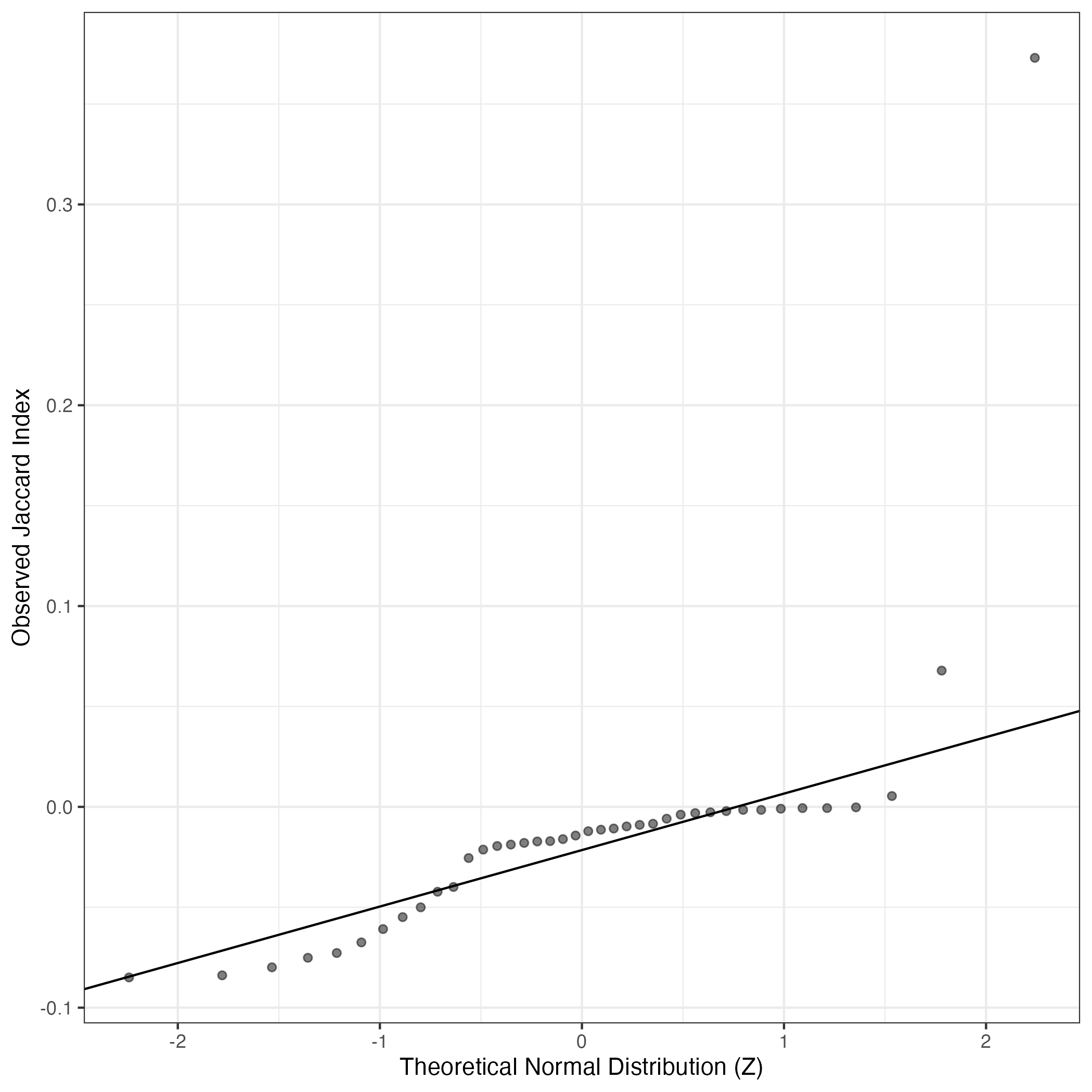
A similar QQ-plot as above, but only for instances where the H value is e.g. > 25, i.e. we are confident that the expression program is active above noise. Agreemenet between these binary vectors is tested using the Jaccard Index, with the P-values calculated by an exact test:
Interactive QQ-plot:
Singler cell type annotations for the top 50 cells on this program.
| Cell ID | Singler label | Singler Delta | Activity Score | Top Singler Raw Scores |
|---|---|---|---|---|
| STDY8004910_TACACGACATGATCCA | Neurons:adrenal_medulla_cell_line | 0.25 | 929.25 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.53, Neuroepithelial_cell:ESC-derived: 0.45, Astrocyte:Embryonic_stem_cell-derived: 0.44, iPS_cells:PDB_1lox-17Puro-10: 0.4, iPS_cells:PDB_1lox-21Puro-20: 0.4, iPS_cells:PDB_1lox-17Puro-5: 0.4, Neurons:ES_cell-derived_neural_precursor: 0.4, iPS_cells:PDB_1lox-21Puro-26: 0.4, iPS_cells:PDB_2lox-5: 0.4, iPS_cells:PDB_2lox-22: 0.4 |
| STDY8004902_TCGCGAGTCTTGTACT | Neurons:adrenal_medulla_cell_line | 0.24 | 832.90 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.51, Neuroepithelial_cell:ESC-derived: 0.43, Astrocyte:Embryonic_stem_cell-derived: 0.43, iPS_cells:PDB_1lox-21Puro-20: 0.4, iPS_cells:PDB_1lox-17Puro-10: 0.4, iPS_cells:PDB_1lox-17Puro-5: 0.4, iPS_cells:PDB_1lox-21Puro-26: 0.4, Neurons:ES_cell-derived_neural_precursor: 0.39, iPS_cells:PDB_2lox-5: 0.39, iPS_cells:PDB_2lox-22: 0.39 |
| STDY8004910_CTCTGGTGTACCCAAT | Neurons:adrenal_medulla_cell_line | 0.24 | 507.70 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.49, Neuroepithelial_cell:ESC-derived: 0.42, Astrocyte:Embryonic_stem_cell-derived: 0.41, iPS_cells:PDB_1lox-21Puro-20: 0.37, iPS_cells:PDB_1lox-17Puro-10: 0.37, Neurons:ES_cell-derived_neural_precursor: 0.37, iPS_cells:PDB_1lox-17Puro-5: 0.37, Embryonic_stem_cells: 0.37, iPS_cells:PDB_1lox-21Puro-26: 0.37, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.37 |
| STDY8004902_CGCGTTTAGCAGCCTC | Neurons:adrenal_medulla_cell_line | 0.24 | 472.36 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.5, Neuroepithelial_cell:ESC-derived: 0.42, Astrocyte:Embryonic_stem_cell-derived: 0.42, Neurons:ES_cell-derived_neural_precursor: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.37, Embryonic_stem_cells: 0.36, iPS_cells:PDB_1lox-17Puro-10: 0.36, iPS_cells:PDB_1lox-17Puro-5: 0.36, iPS_cells:PDB_1lox-21Puro-26: 0.36, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.36 |
| STDY8004902_TTGCCGTCAGTATGCT | Neurons:adrenal_medulla_cell_line | 0.23 | 471.24 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.49, Neuroepithelial_cell:ESC-derived: 0.42, Astrocyte:Embryonic_stem_cell-derived: 0.41, iPS_cells:PDB_1lox-21Puro-20: 0.38, iPS_cells:PDB_1lox-17Puro-10: 0.38, iPS_cells:PDB_1lox-17Puro-5: 0.37, Neurons:ES_cell-derived_neural_precursor: 0.37, iPS_cells:PDB_1lox-21Puro-26: 0.37, iPS_cells:PDB_2lox-22: 0.37, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.37 |
| STDY8004910_CCAGCGACACACAGAG | Neurons:adrenal_medulla_cell_line | 0.21 | 465.35 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.43, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.37, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, Neurons:ES_cell-derived_neural_precursor: 0.34, iPS_cells:PDB_2lox-22: 0.34, iPS_cells:PDB_2lox-5: 0.34 |
| STDY8004902_GGTGCGTAGGACTGGT | Neurons:adrenal_medulla_cell_line | 0.24 | 437.78 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.51, Neuroepithelial_cell:ESC-derived: 0.44, Astrocyte:Embryonic_stem_cell-derived: 0.43, iPS_cells:PDB_1lox-21Puro-20: 0.4, iPS_cells:PDB_1lox-17Puro-10: 0.4, iPS_cells:PDB_1lox-17Puro-5: 0.39, iPS_cells:PDB_1lox-21Puro-26: 0.39, Neurons:ES_cell-derived_neural_precursor: 0.39, iPS_cells:PDB_2lox-22: 0.39, Embryonic_stem_cells: 0.39 |
| STDY8004910_ATTCTACAGAACTCGG | Neurons:adrenal_medulla_cell_line | 0.24 | 424.81 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.5, Neuroepithelial_cell:ESC-derived: 0.42, Astrocyte:Embryonic_stem_cell-derived: 0.42, iPS_cells:PDB_1lox-21Puro-20: 0.38, iPS_cells:PDB_1lox-17Puro-10: 0.37, Neurons:ES_cell-derived_neural_precursor: 0.37, Embryonic_stem_cells: 0.37, iPS_cells:PDB_1lox-17Puro-5: 0.37, iPS_cells:PDB_2lox-22: 0.37, iPS_cells:PDB_1lox-21Puro-26: 0.37 |
| STDY8004902_CGAGCACTCTTCATGT | Neurons:adrenal_medulla_cell_line | 0.21 | 414.11 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.48, Neuroepithelial_cell:ESC-derived: 0.41, Astrocyte:Embryonic_stem_cell-derived: 0.4, iPS_cells:PDB_1lox-21Puro-20: 0.37, iPS_cells:PDB_1lox-17Puro-10: 0.37, iPS_cells:PDB_1lox-17Puro-5: 0.37, iPS_cells:PDB_2lox-22: 0.37, iPS_cells:PDB_1lox-21Puro-26: 0.37, iPS_cells:PDB_2lox-5: 0.37, iPS_cells:PDB_2lox-17: 0.37 |
| STDY8004910_CAGCCGAGTGTGAATA | Neurons:adrenal_medulla_cell_line | 0.23 | 394.04 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.48, Neuroepithelial_cell:ESC-derived: 0.41, Astrocyte:Embryonic_stem_cell-derived: 0.4, Neurons:ES_cell-derived_neural_precursor: 0.36, iPS_cells:PDB_1lox-17Puro-10: 0.36, iPS_cells:PDB_1lox-21Puro-20: 0.36, iPS_cells:PDB_1lox-17Puro-5: 0.36, iPS_cells:PDB_2lox-22: 0.36, Embryonic_stem_cells: 0.36, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.35 |
| STDY8004910_CAACCAAGTTCCACGG | Neurons:adrenal_medulla_cell_line | 0.24 | 364.13 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.47, Neuroepithelial_cell:ESC-derived: 0.41, Astrocyte:Embryonic_stem_cell-derived: 0.4, Neurons:ES_cell-derived_neural_precursor: 0.36, iPS_cells:PDB_1lox-21Puro-20: 0.36, iPS_cells:PDB_1lox-17Puro-10: 0.36, iPS_cells:PDB_1lox-17Puro-5: 0.36, iPS_cells:PDB_1lox-21Puro-26: 0.35, iPS_cells:PDB_2lox-22: 0.35, Embryonic_stem_cells: 0.35 |
| STDY8004902_GCACTCTAGTTCGATC | Neurons:adrenal_medulla_cell_line | 0.26 | 356.10 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.52, Neuroepithelial_cell:ESC-derived: 0.44, Astrocyte:Embryonic_stem_cell-derived: 0.43, iPS_cells:PDB_1lox-21Puro-20: 0.41, Neurons:ES_cell-derived_neural_precursor: 0.4, iPS_cells:PDB_1lox-17Puro-10: 0.4, iPS_cells:PDB_1lox-21Puro-26: 0.4, iPS_cells:PDB_1lox-17Puro-5: 0.4, iPS_cells:PDB_2lox-22: 0.4, iPS_cells:PDB_2lox-5: 0.39 |
| STDY8004894_AGGGATGTCAGCTTAG | Neurons:adrenal_medulla_cell_line | 0.14 | 355.99 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.42, Neuroepithelial_cell:ESC-derived: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.35, Neurons:Schwann_cell: 0.32, iPS_cells:PDB_1lox-21Puro-20: 0.31, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.31, iPS_cells:PDB_1lox-17Puro-10: 0.31, iPS_cells:PDB_1lox-17Puro-5: 0.31, Neurons:ES_cell-derived_neural_precursor: 0.31, iPS_cells:PDB_1lox-21Puro-26: 0.31 |
| STDY8004902_CATGACATCTACTATC | Neurons:adrenal_medulla_cell_line | 0.23 | 345.84 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.46, Neuroepithelial_cell:ESC-derived: 0.4, Astrocyte:Embryonic_stem_cell-derived: 0.39, Neurons:ES_cell-derived_neural_precursor: 0.36, iPS_cells:PDB_1lox-21Puro-20: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.35, iPS_cells:PDB_1lox-21Puro-26: 0.35, Embryonic_stem_cells: 0.35, iPS_cells:PDB_2lox-22: 0.35 |
| STDY8004902_TGTTCCGTCAGCGACC | Neurons:adrenal_medulla_cell_line | 0.20 | 332.11 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.46, Neuroepithelial_cell:ESC-derived: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.35, iPS_cells:PDB_1lox-21Puro-26: 0.35, Neurons:ES_cell-derived_neural_precursor: 0.35, iPS_cells:PDB_2lox-22: 0.35, Embryonic_stem_cells: 0.35 |
| STDY8004902_CCTACACAGTCAAGGC | Neurons:adrenal_medulla_cell_line | 0.22 | 325.57 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.47, Neuroepithelial_cell:ESC-derived: 0.41, Astrocyte:Embryonic_stem_cell-derived: 0.39, iPS_cells:PDB_1lox-17Puro-10: 0.37, iPS_cells:PDB_1lox-21Puro-20: 0.37, iPS_cells:PDB_1lox-17Puro-5: 0.36, iPS_cells:PDB_2lox-22: 0.36, iPS_cells:PDB_1lox-21Puro-26: 0.36, Neurons:ES_cell-derived_neural_precursor: 0.36, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.36 |
| STDY8004902_GGCAATTCAGGGCATA | Neurons:adrenal_medulla_cell_line | 0.22 | 323.45 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.46, Neuroepithelial_cell:ESC-derived: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_2lox-22: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.35, iPS_cells:PDB_2lox-5: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.35, iPS_cells:PDB_1lox-21Puro-26: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.35, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.35 |
| STDY8004910_AGGTCCGTCAGTACGT | Neurons:adrenal_medulla_cell_line | 0.21 | 322.89 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.43, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.36, Neurons:ES_cell-derived_neural_precursor: 0.34, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.33, iPS_cells:PDB_2lox-22: 0.33, iPS_cells:PDB_1lox-21Puro-26: 0.33, Embryonic_stem_cells: 0.33 |
| STDY8004910_CCGTTCAAGGAGTACC | Neurons:adrenal_medulla_cell_line | 0.21 | 322.01 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.45, Neuroepithelial_cell:ESC-derived: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.35, Neurons:ES_cell-derived_neural_precursor: 0.35, iPS_cells:PDB_1lox-21Puro-26: 0.35, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.35, Embryonic_stem_cells: 0.35 |
| STDY8004910_CACCACTCAGACAAGC | Neurons:adrenal_medulla_cell_line | 0.25 | 320.14 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.54, Neuroepithelial_cell:ESC-derived: 0.47, Astrocyte:Embryonic_stem_cell-derived: 0.45, iPS_cells:PDB_1lox-21Puro-20: 0.43, iPS_cells:PDB_1lox-21Puro-26: 0.43, iPS_cells:PDB_1lox-17Puro-10: 0.43, iPS_cells:PDB_2lox-22: 0.43, iPS_cells:PDB_1lox-17Puro-5: 0.43, Neurons:ES_cell-derived_neural_precursor: 0.42, iPS_cells:PDB_2lox-17: 0.42 |
| STDY8004902_CAGCCGATCACAACGT | Neurons:adrenal_medulla_cell_line | 0.21 | 317.40 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.45, Neuroepithelial_cell:ESC-derived: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.38, Neurons:ES_cell-derived_neural_precursor: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.34, Embryonic_stem_cells: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_2lox-22: 0.34 |
| STDY8004902_ACTTACTTCTTAGAGC | Neurons:adrenal_medulla_cell_line | 0.25 | 309.13 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.5, Neuroepithelial_cell:ESC-derived: 0.44, Astrocyte:Embryonic_stem_cell-derived: 0.42, iPS_cells:PDB_1lox-21Puro-20: 0.41, iPS_cells:PDB_1lox-17Puro-10: 0.41, iPS_cells:PDB_1lox-17Puro-5: 0.41, iPS_cells:PDB_1lox-21Puro-26: 0.4, iPS_cells:PDB_2lox-22: 0.4, Embryonic_stem_cells: 0.4, iPS_cells:PDB_2lox-21: 0.4 |
| STDY8004902_CTCCTAGTCAGTCCCT | Neurons:adrenal_medulla_cell_line | 0.22 | 304.63 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.42, Neuroepithelial_cell:ESC-derived: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.34, Neurons:ES_cell-derived_neural_precursor: 0.31, iPS_cells:PDB_1lox-17Puro-10: 0.3, iPS_cells:PDB_1lox-21Puro-20: 0.3, iPS_cells:PDB_1lox-21Puro-26: 0.3, iPS_cells:PDB_1lox-17Puro-5: 0.3, Embryonic_stem_cells: 0.3, iPS_cells:PDB_2lox-22: 0.3 |
| STDY8004910_CAGGTGCAGCGTCTAT | Neurons:adrenal_medulla_cell_line | 0.20 | 297.96 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.42, Neuroepithelial_cell:ESC-derived: 0.37, Astrocyte:Embryonic_stem_cell-derived: 0.36, iPS_cells:PDB_1lox-21Puro-20: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.32, iPS_cells:PDB_1lox-21Puro-26: 0.32, iPS_cells:PDB_1lox-17Puro-10: 0.32, Neurons:ES_cell-derived_neural_precursor: 0.32, Embryonic_stem_cells: 0.32, iPS_cells:PDB_2lox-22: 0.32 |
| STDY8004910_TACAGTGTCGGTCTAA | Neurons:adrenal_medulla_cell_line | 0.20 | 296.87 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.42, Astrocyte:Embryonic_stem_cell-derived: 0.37, Neuroepithelial_cell:ESC-derived: 0.37, Neurons:ES_cell-derived_neural_precursor: 0.32, Neurons:Schwann_cell: 0.32, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.32, iPS_cells:PDB_1lox-17Puro-10: 0.32, Embryonic_stem_cells: 0.32, iPS_cells:PDB_1lox-17Puro-5: 0.32, iPS_cells:PDB_1lox-21Puro-20: 0.32 |
| STDY8004902_CAGCGACTCGCGGATC | Neurons:adrenal_medulla_cell_line | 0.23 | 294.33 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.48, Astrocyte:Embryonic_stem_cell-derived: 0.4, Neuroepithelial_cell:ESC-derived: 0.4, iPS_cells:PDB_1lox-21Puro-20: 0.36, iPS_cells:PDB_1lox-17Puro-10: 0.36, iPS_cells:PDB_1lox-17Puro-5: 0.36, iPS_cells:PDB_2lox-22: 0.36, iPS_cells:PDB_1lox-21Puro-26: 0.36, iPS_cells:iPS:minicircle-derived: 0.35, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.35 |
| STDY8004902_TCATTTGCATAAAGGT | Neurons:adrenal_medulla_cell_line | 0.20 | 293.49 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.46, Neuroepithelial_cell:ESC-derived: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.35, Embryonic_stem_cells: 0.35, iPS_cells:PDB_1lox-21Puro-26: 0.35, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.35, Neurons:ES_cell-derived_neural_precursor: 0.35 |
| STDY8004910_GAGTCCGCAGAGCCAA | Neurons:adrenal_medulla_cell_line | 0.22 | 285.97 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.45, Neuroepithelial_cell:ESC-derived: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.38, Neurons:ES_cell-derived_neural_precursor: 0.33, iPS_cells:PDB_1lox-21Puro-20: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.33, Embryonic_stem_cells: 0.33, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.33, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.33 |
| STDY8004910_CGTAGGCTCTCGTTTA | Neurons:adrenal_medulla_cell_line | 0.21 | 283.70 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.44, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.37, Neurons:ES_cell-derived_neural_precursor: 0.34, iPS_cells:PDB_1lox-21Puro-20: 0.34, Embryonic_stem_cells: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.33, iPS_cells:PDB_2lox-22: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.33, iPS_cells:PDB_1lox-21Puro-26: 0.33 |
| STDY8004902_CGTCTACAGGTTCCTA | Neurons:adrenal_medulla_cell_line | 0.23 | 282.62 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.46, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.37, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.34, iPS_cells:PDB_2lox-22: 0.34, Neurons:ES_cell-derived_neural_precursor: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_2lox-5: 0.34 |
| STDY8004902_TGAGCCGCACCGCTAG | Neurons:adrenal_medulla_cell_line | 0.21 | 281.74 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.47, Neuroepithelial_cell:ESC-derived: 0.4, Astrocyte:Embryonic_stem_cell-derived: 0.39, iPS_cells:PDB_1lox-21Puro-20: 0.36, iPS_cells:iPS:minicircle-derived: 0.36, iPS_cells:PDB_1lox-17Puro-10: 0.36, Neurons:ES_cell-derived_neural_precursor: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.35, iPS_cells:skin_fibroblast-derived: 0.35, iPS_cells:PDB_1lox-21Puro-26: 0.35 |
| STDY8004910_TGCTACCCACGGCTAC | Neurons:adrenal_medulla_cell_line | 0.23 | 278.29 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.47, Neuroepithelial_cell:ESC-derived: 0.4, Astrocyte:Embryonic_stem_cell-derived: 0.4, iPS_cells:PDB_1lox-21Puro-20: 0.35, Neurons:ES_cell-derived_neural_precursor: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.35, iPS_cells:PDB_1lox-21Puro-26: 0.35, Embryonic_stem_cells: 0.35, iPS_cells:PDB_2lox-22: 0.35 |
| STDY8004902_TAGTGGTAGGCTACGA | Neurons:adrenal_medulla_cell_line | 0.22 | 275.22 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.44, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.37, Neurons:ES_cell-derived_neural_precursor: 0.34, iPS_cells:PDB_1lox-21Puro-20: 0.33, Embryonic_stem_cells: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.33, iPS_cells:PDB_1lox-21Puro-26: 0.33, iPS_cells:PDB_2lox-22: 0.32 |
| STDY8004910_TCAGATGTCAGTCAGT | Neurons:adrenal_medulla_cell_line | 0.20 | 274.89 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.44, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.37, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.33, iPS_cells:PDB_1lox-17Puro-5: 0.33, iPS_cells:PDB_2lox-22: 0.33, iPS_cells:PDB_1lox-21Puro-26: 0.33, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.33, Embryonic_stem_cells: 0.33 |
| STDY8004902_TCTGGAAGTAAACGCG | Neurons:adrenal_medulla_cell_line | 0.21 | 274.55 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.46, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_2lox-22: 0.34, Neurons:ES_cell-derived_neural_precursor: 0.34, iPS_cells:PDB_2lox-21: 0.34 |
| STDY8004902_GACGCGTTCACATAGC | Neurons:adrenal_medulla_cell_line | 0.20 | 271.20 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.41, Neuroepithelial_cell:ESC-derived: 0.36, Astrocyte:Embryonic_stem_cell-derived: 0.35, Neurons:ES_cell-derived_neural_precursor: 0.32, iPS_cells:PDB_1lox-21Puro-20: 0.31, iPS_cells:PDB_1lox-17Puro-10: 0.31, iPS_cells:PDB_1lox-17Puro-5: 0.31, Embryonic_stem_cells: 0.31, iPS_cells:PDB_1lox-21Puro-26: 0.31, iPS_cells:PDB_2lox-22: 0.31 |
| STDY8004910_CTCTACGTCATACGGT | Neurons:adrenal_medulla_cell_line | 0.20 | 269.63 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.42, Astrocyte:Embryonic_stem_cell-derived: 0.36, Neuroepithelial_cell:ESC-derived: 0.36, iPS_cells:PDB_1lox-21Puro-20: 0.32, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.32, Neurons:ES_cell-derived_neural_precursor: 0.32, Embryonic_stem_cells: 0.32, iPS_cells:PDB_2lox-22: 0.31, iPS_cells:PDB_1lox-21Puro-26: 0.31, iPS_cells:iPS:minicircle-derived: 0.31 |
| STDY8004894_AAACGGGCAGTAACGG | Neurons:adrenal_medulla_cell_line | 0.13 | 268.67 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.41, Neuroepithelial_cell:ESC-derived: 0.35, Astrocyte:Embryonic_stem_cell-derived: 0.35, iPS_cells:PDB_2lox-22: 0.32, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.32, iPS_cells:PDB_1lox-21Puro-20: 0.32, iPS_cells:PDB_2lox-17: 0.32, iPS_cells:PDB_1lox-17Puro-5: 0.32, iPS_cells:iPS:minicircle-derived: 0.32, iPS_cells:PDB_2lox-21: 0.32 |
| STDY8004910_GGTGCGTCATATGGTC | Neurons:adrenal_medulla_cell_line | 0.21 | 261.76 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.45, Neuroepithelial_cell:ESC-derived: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.36, iPS_cells:PDB_1lox-17Puro-10: 0.36, iPS_cells:PDB_1lox-21Puro-26: 0.35, iPS_cells:PDB_2lox-22: 0.35, Neurons:ES_cell-derived_neural_precursor: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.35, Embryonic_stem_cells: 0.35 |
| STDY8004902_GGAACTTAGTGGCACA | Neurons:adrenal_medulla_cell_line | 0.18 | 261.28 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.44, Neuroepithelial_cell:ESC-derived: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:iPS:minicircle-derived: 0.36, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.36, iPS_cells:PDB_1lox-17Puro-5: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.35, iPS_cells:skin_fibroblast-derived: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.35, Embryonic_stem_cells: 0.35 |
| STDY8004902_GCTTGAAAGCAGATCG | Neurons:adrenal_medulla_cell_line | 0.21 | 260.85 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.47, Neuroepithelial_cell:ESC-derived: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.35, Embryonic_stem_cells: 0.35, iPS_cells:PDB_1lox-17Puro-10: 0.35, iPS_cells:PDB_1lox-17Puro-5: 0.35, iPS_cells:PDB_2lox-22: 0.35, Neurons:ES_cell-derived_neural_precursor: 0.35, iPS_cells:PDB_1lox-21Puro-26: 0.35 |
| STDY8004910_CGTCACTTCATAACCG | Neurons:adrenal_medulla_cell_line | 0.20 | 259.04 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.43, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.37, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_2lox-22: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, Neurons:ES_cell-derived_neural_precursor: 0.34, Embryonic_stem_cells: 0.34 |
| STDY8004902_CCTACACAGCCACTAT | Neurons:adrenal_medulla_cell_line | 0.23 | 253.62 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.45, Neuroepithelial_cell:ESC-derived: 0.38, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.34, iPS_cells:PDB_1lox-17Puro-10: 0.34, iPS_cells:PDB_1lox-21Puro-26: 0.34, iPS_cells:PDB_1lox-17Puro-5: 0.34, Neurons:ES_cell-derived_neural_precursor: 0.34, iPS_cells:PDB_2lox-22: 0.34, Embryonic_stem_cells: 0.33 |
| STDY8004902_CTCCTAGGTTACGGAG | Neurons:adrenal_medulla_cell_line | 0.23 | 252.04 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.48, Neuroepithelial_cell:ESC-derived: 0.41, Astrocyte:Embryonic_stem_cell-derived: 0.4, iPS_cells:PDB_1lox-21Puro-20: 0.37, iPS_cells:PDB_1lox-17Puro-10: 0.37, Neurons:ES_cell-derived_neural_precursor: 0.37, iPS_cells:PDB_1lox-17Puro-5: 0.37, iPS_cells:PDB_1lox-21Puro-26: 0.37, iPS_cells:PDB_2lox-22: 0.37, iPS_cells:PDB_2lox-5: 0.36 |
| STDY8004910_AGCGTATAGTTCGATC | Neurons:adrenal_medulla_cell_line | 0.20 | 247.05 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.44, Neuroepithelial_cell:ESC-derived: 0.39, Astrocyte:Embryonic_stem_cell-derived: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.36, iPS_cells:PDB_1lox-17Puro-10: 0.36, iPS_cells:PDB_1lox-21Puro-26: 0.36, iPS_cells:PDB_1lox-17Puro-5: 0.36, iPS_cells:PDB_2lox-22: 0.35, Neurons:ES_cell-derived_neural_precursor: 0.35, Embryonic_stem_cells: 0.35 |
| STDY8004910_TACAGTGGTTACCGAT | Neurons:adrenal_medulla_cell_line | 0.20 | 244.76 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.41, Neuroepithelial_cell:ESC-derived: 0.36, Astrocyte:Embryonic_stem_cell-derived: 0.35, iPS_cells:PDB_1lox-21Puro-20: 0.32, iPS_cells:PDB_1lox-17Puro-10: 0.32, iPS_cells:PDB_1lox-17Puro-5: 0.32, Neurons:ES_cell-derived_neural_precursor: 0.32, iPS_cells:PDB_2lox-22: 0.32, iPS_cells:PDB_1lox-21Puro-26: 0.32, Embryonic_stem_cells: 0.32 |
| STDY8004902_CGATCGGAGACAAGCC | Neurons:adrenal_medulla_cell_line | 0.25 | 243.61 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.5, Neuroepithelial_cell:ESC-derived: 0.43, Astrocyte:Embryonic_stem_cell-derived: 0.4, Neurons:ES_cell-derived_neural_precursor: 0.38, iPS_cells:PDB_1lox-21Puro-20: 0.38, iPS_cells:PDB_1lox-21Puro-26: 0.38, iPS_cells:PDB_1lox-17Puro-5: 0.38, iPS_cells:PDB_1lox-17Puro-10: 0.38, Embryonic_stem_cells: 0.38, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.38 |
| STDY8004902_GATCTAGAGATGTGTA | Neurons:adrenal_medulla_cell_line | 0.21 | 241.20 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.45, Astrocyte:Embryonic_stem_cell-derived: 0.37, Neuroepithelial_cell:ESC-derived: 0.37, Neurons:ES_cell-derived_neural_precursor: 0.34, iPS_cells:PDB_1lox-21Puro-20: 0.33, iPS_cells:PDB_1lox-17Puro-10: 0.33, iPS_cells:PDB_1lox-21Puro-26: 0.33, iPS_cells:PDB_2lox-22: 0.33, Embryonic_stem_cells: 0.33, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.33 |
| STDY8004910_ATAACGCTCATCGATG | Neurons:adrenal_medulla_cell_line | 0.19 | 240.61 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.43, Neuroepithelial_cell:ESC-derived: 0.37, Astrocyte:Embryonic_stem_cell-derived: 0.36, Embryonic_stem_cells: 0.32, iPS_cells:PDB_1lox-21Puro-20: 0.32, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.32, iPS_cells:PDB_1lox-17Puro-10: 0.32, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.32, iPS_cells:PDB_1lox-17Puro-5: 0.32, iPS_cells:PDB_1lox-21Puro-26: 0.32 |
| STDY8004902_GGACGTCCACAGGCCT | Neurons:adrenal_medulla_cell_line | 0.24 | 233.05 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.5, Neuroepithelial_cell:ESC-derived: 0.44, Astrocyte:Embryonic_stem_cell-derived: 0.42, iPS_cells:PDB_1lox-21Puro-20: 0.4, iPS_cells:PDB_1lox-17Puro-10: 0.4, iPS_cells:PDB_1lox-17Puro-5: 0.4, iPS_cells:PDB_1lox-21Puro-26: 0.4, iPS_cells:PDB_2lox-22: 0.4, Neurons:ES_cell-derived_neural_precursor: 0.39, Embryonic_stem_cells: 0.39 |
Below shows the significant enrichments of this GEP for literature curated gene lists
This data was procured from existing single cell RNA-seq maps of neuroblastoma or related relevant data.
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by the P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Medulla (Hanemaaijer)
Marker genes obtained from Supplementary Table SD of Hanemaaijer et al (PMID 33500353). The authors generated single-cell RNA-seq data (sort-seq, 2,229 cells total) from mouse adrenal glads at E13.5, E14.5, E17.5, E18.5, P1 and P5. These were marker genes that matched with a similar dataset generated by Furlan et al (PMID 28684471). This particular set of markers are for genes defining the broad group labelled adrenal medulla, which included 7 subclusters (SCP, Bridge, Committed Progenitor, N Chromaffin, E Chromaffin, Neuroblast) - see UMAP on their Fig1B for cluster assignments.:
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.49e-11
Mean rank of genes in gene set: 258.12
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TH | 0.0087457 | 6 | GTEx | DepMap | Descartes | 5.54 | 940.02 |
| DBH | 0.0085132 | 8 | GTEx | DepMap | Descartes | 6.71 | 778.96 |
| PHOX2A | 0.0074979 | 15 | GTEx | DepMap | Descartes | 5.18 | 888.99 |
| CHGA | 0.0072050 | 16 | GTEx | DepMap | Descartes | 4.25 | 662.30 |
| UCHL1 | 0.0070231 | 19 | GTEx | DepMap | Descartes | 12.20 | 2291.55 |
| PCSK1N | 0.0067364 | 23 | GTEx | DepMap | Descartes | 14.49 | 4392.91 |
| DDC | 0.0062103 | 32 | GTEx | DepMap | Descartes | 2.60 | 384.59 |
| MAP1B | 0.0058948 | 41 | GTEx | DepMap | Descartes | 24.19 | 598.30 |
| SLC18A1 | 0.0056771 | 48 | GTEx | DepMap | Descartes | 1.28 | 137.40 |
| CYB561 | 0.0052510 | 66 | GTEx | DepMap | Descartes | 1.02 | 102.11 |
| CHGB | 0.0041548 | 174 | GTEx | DepMap | Descartes | 12.23 | 1517.14 |
| GATA3 | 0.0039683 | 199 | GTEx | DepMap | Descartes | 4.59 | 467.86 |
| HAND2 | 0.0036954 | 241 | GTEx | DepMap | Descartes | 4.46 | 497.57 |
| EML5 | 0.0026840 | 495 | GTEx | DepMap | Descartes | 0.46 | 14.70 |
| NNAT | 0.0016179 | 1188 | GTEx | DepMap | Descartes | 12.58 | 2911.00 |
| DISP2 | 0.0013014 | 1559 | GTEx | DepMap | Descartes | 0.39 | 9.84 |
Adrenergic Fig 1D (Olsen)
Similar to above, but for selected adrenergic marker genes:
Wilcoxon ranksum test P-value for gene set overrepresentation: 4.89e-08
Mean rank of genes in gene set: 454.91
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| RGS5 | 0.0106499 | 1 | GTEx | DepMap | Descartes | 17.67 | 940.52 |
| TH | 0.0087457 | 6 | GTEx | DepMap | Descartes | 5.54 | 940.02 |
| DBH | 0.0085132 | 8 | GTEx | DepMap | Descartes | 6.71 | 778.96 |
| ELAVL4 | 0.0084525 | 9 | GTEx | DepMap | Descartes | 11.34 | 845.99 |
| MLLT11 | 0.0077011 | 14 | GTEx | DepMap | Descartes | 27.14 | 3321.19 |
| UCHL1 | 0.0070231 | 19 | GTEx | DepMap | Descartes | 12.20 | 2291.55 |
| STMN2 | 0.0067652 | 22 | GTEx | DepMap | Descartes | 70.70 | 10962.20 |
| MAP1B | 0.0058948 | 41 | GTEx | DepMap | Descartes | 24.19 | 598.30 |
| ISL1 | 0.0035492 | 266 | GTEx | DepMap | Descartes | 2.36 | 307.91 |
| RTN1 | 0.0025415 | 561 | GTEx | DepMap | Descartes | 29.66 | 2865.34 |
| NRG1 | 0.0003726 | 4057 | GTEx | DepMap | Descartes | 0.31 | 6.90 |
N Chromafin (Hanemaaijer)
Marker genes obtained from Supplementary Table SD of Hanemaaijer et al (PMID 33500353). The authors generated single-cell RNA-seq data (sort-seq, 2,229 cells total) from mouse adrenal glads at E13.5, E14.5, E17.5, E18.5, P1 and P5. These were marker genes that matched with a similar dataset generated by Furlan et al (PMID 28684471). This particular set of markers are for the Norepinepherine Chromaffin subcluster, which is part of the Adrenal Medulla cluster.:
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.17e-06
Mean rank of genes in gene set: 2116.62
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| GNAS | 0.0071699 | 17 | GTEx | DepMap | Descartes | 25.92 | 2033.26 |
| C1QL1 | 0.0068111 | 21 | GTEx | DepMap | Descartes | 1.56 | 283.35 |
| SCG5 | 0.0061786 | 33 | GTEx | DepMap | Descartes | 3.29 | 853.39 |
| SNAP25 | 0.0056728 | 49 | GTEx | DepMap | Descartes | 3.93 | 525.20 |
| SCG3 | 0.0051254 | 76 | GTEx | DepMap | Descartes | 2.73 | 257.86 |
| CELF4 | 0.0047708 | 110 | GTEx | DepMap | Descartes | 3.85 | 312.10 |
| PTPRN | 0.0041202 | 181 | GTEx | DepMap | Descartes | 1.06 | 63.42 |
| SLCO3A1 | 0.0038250 | 216 | GTEx | DepMap | Descartes | 1.89 | 117.05 |
| NAP1L5 | 0.0028754 | 428 | GTEx | DepMap | Descartes | 1.06 | 168.47 |
| PCLO | 0.0025291 | 569 | GTEx | DepMap | Descartes | 1.61 | 25.70 |
| CACNA2D1 | 0.0009821 | 2085 | GTEx | DepMap | Descartes | 0.67 | 27.37 |
| PPFIA2 | 0.0008751 | 2346 | GTEx | DepMap | Descartes | 0.19 | 10.47 |
| SYN2 | 0.0007518 | 2639 | GTEx | DepMap | Descartes | 0.08 | 5.40 |
| LGR5 | -0.0001620 | 7946 | GTEx | DepMap | Descartes | 0.00 | 0.53 |
| ADCYAP1R1 | -0.0001645 | 7977 | GTEx | DepMap | Descartes | 0.06 | 2.27 |
| CXCL14 | -0.0002864 | 9173 | GTEx | DepMap | Descartes | 0.06 | 8.65 |
Below shows ranks on this GEP for literature curated gene lists for large gene sets
These include those reported as mesenchymal/adrenergic by Van Groningen et al.
High ranks indicate this gene is a driver of this GEP (note these results are not ordered).
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
VanGroningen Adrenergic Genes
Adrenergic marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 5.71e-48
Mean rank of genes in gene set: 3303.11
Median rank of genes in gene set: 1483
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| RGS5 | 0.0106499 | 1 | GTEx | DepMap | Descartes | 17.67 | 940.52 |
| TH | 0.0087457 | 6 | GTEx | DepMap | Descartes | 5.54 | 940.02 |
| DBH | 0.0085132 | 8 | GTEx | DepMap | Descartes | 6.71 | 778.96 |
| ELAVL4 | 0.0084525 | 9 | GTEx | DepMap | Descartes | 11.34 | 845.99 |
| PHOX2A | 0.0074979 | 15 | GTEx | DepMap | Descartes | 5.18 | 888.99 |
| CHGA | 0.0072050 | 16 | GTEx | DepMap | Descartes | 4.25 | 662.30 |
| TMOD1 | 0.0070401 | 18 | GTEx | DepMap | Descartes | 5.07 | 491.72 |
| STMN2 | 0.0067652 | 22 | GTEx | DepMap | Descartes | 70.70 | 10962.20 |
| TUBB2B | 0.0064973 | 29 | GTEx | DepMap | Descartes | 28.88 | 4404.20 |
| DNER | 0.0062353 | 31 | GTEx | DepMap | Descartes | 1.34 | 137.58 |
| DDC | 0.0062103 | 32 | GTEx | DepMap | Descartes | 2.60 | 384.59 |
| GAP43 | 0.0060681 | 35 | GTEx | DepMap | Descartes | 16.43 | 2557.78 |
| TUBB2A | 0.0060213 | 37 | GTEx | DepMap | Descartes | 19.74 | 3476.18 |
| MAP1B | 0.0058948 | 41 | GTEx | DepMap | Descartes | 24.19 | 598.30 |
| QDPR | 0.0058466 | 42 | GTEx | DepMap | Descartes | 3.34 | 657.20 |
| NRCAM | 0.0057528 | 45 | GTEx | DepMap | Descartes | 1.36 | 71.47 |
| SNAP25 | 0.0056728 | 49 | GTEx | DepMap | Descartes | 3.93 | 525.20 |
| TAGLN3 | 0.0056168 | 51 | GTEx | DepMap | Descartes | 2.73 | 559.68 |
| ACOT7 | 0.0055337 | 55 | GTEx | DepMap | Descartes | 3.38 | 399.11 |
| RUNDC3B | 0.0052542 | 65 | GTEx | DepMap | Descartes | 1.16 | 90.14 |
| FKBP1B | 0.0051994 | 70 | GTEx | DepMap | Descartes | 1.72 | 321.53 |
| GATA2 | 0.0051803 | 73 | GTEx | DepMap | Descartes | 1.76 | 166.14 |
| SYT1 | 0.0051797 | 74 | GTEx | DepMap | Descartes | 3.62 | 224.53 |
| SCG3 | 0.0051254 | 76 | GTEx | DepMap | Descartes | 2.73 | 257.86 |
| MMD | 0.0051252 | 77 | GTEx | DepMap | Descartes | 3.08 | 340.29 |
| PHYHIPL | 0.0050616 | 79 | GTEx | DepMap | Descartes | 0.75 | 60.12 |
| SYT4 | 0.0050487 | 81 | GTEx | DepMap | Descartes | 1.76 | 137.18 |
| RUNDC3A | 0.0050277 | 83 | GTEx | DepMap | Descartes | 1.95 | 160.43 |
| HAND1 | 0.0049860 | 89 | GTEx | DepMap | Descartes | 1.37 | 242.56 |
| TSPAN7 | 0.0049077 | 93 | GTEx | DepMap | Descartes | 2.26 | 397.61 |
VanGroningen Mesenchymal Genes
Mesenchymal marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 8625.9
Median rank of genes in gene set: 10171
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ATP1B1 | 0.0051829 | 72 | GTEx | DepMap | Descartes | 4.80 | 608.32 |
| GNAI1 | 0.0048198 | 104 | GTEx | DepMap | Descartes | 1.18 | 37.64 |
| ANXA2 | 0.0046228 | 123 | GTEx | DepMap | Descartes | 9.96 | 868.93 |
| DLC1 | 0.0036047 | 255 | GTEx | DepMap | Descartes | 1.55 | 72.40 |
| TMEFF2 | 0.0035797 | 260 | GTEx | DepMap | Descartes | 1.88 | 171.88 |
| NES | 0.0034739 | 289 | GTEx | DepMap | Descartes | 1.24 | 76.25 |
| FAM3C | 0.0033214 | 312 | GTEx | DepMap | Descartes | 1.85 | 240.85 |
| CBFB | 0.0033199 | 314 | GTEx | DepMap | Descartes | 1.81 | 189.53 |
| CRABP2 | 0.0028797 | 423 | GTEx | DepMap | Descartes | 1.08 | 318.80 |
| SHC1 | 0.0026261 | 523 | GTEx | DepMap | Descartes | 1.34 | 118.46 |
| TPM1 | 0.0025583 | 549 | GTEx | DepMap | Descartes | 2.26 | 169.23 |
| ARMCX2 | 0.0024862 | 592 | GTEx | DepMap | Descartes | 0.42 | 49.75 |
| RHOC | 0.0024804 | 597 | GTEx | DepMap | Descartes | 2.48 | 345.88 |
| CTNNA1 | 0.0024578 | 609 | GTEx | DepMap | Descartes | 1.00 | 82.49 |
| SERPINE2 | 0.0023426 | 655 | GTEx | DepMap | Descartes | 1.68 | 81.20 |
| ITM2C | 0.0022153 | 717 | GTEx | DepMap | Descartes | 2.67 | 437.89 |
| LAPTM4A | 0.0019448 | 895 | GTEx | DepMap | Descartes | 2.94 | 668.85 |
| ATP2B4 | 0.0019298 | 915 | GTEx | DepMap | Descartes | 0.86 | 30.70 |
| PLAGL1 | 0.0018561 | 975 | GTEx | DepMap | Descartes | 0.65 | 35.32 |
| RGS3 | 0.0017388 | 1055 | GTEx | DepMap | Descartes | 0.43 | 27.35 |
| SDC2 | 0.0015902 | 1212 | GTEx | DepMap | Descartes | 0.53 | 52.86 |
| PDIA3 | 0.0015775 | 1223 | GTEx | DepMap | Descartes | 2.95 | 242.34 |
| SCPEP1 | 0.0015675 | 1233 | GTEx | DepMap | Descartes | 0.88 | 161.16 |
| DKK3 | 0.0014848 | 1334 | GTEx | DepMap | Descartes | 0.50 | 16.05 |
| ENAH | 0.0014134 | 1422 | GTEx | DepMap | Descartes | 1.21 | 26.67 |
| NRP1 | 0.0013377 | 1509 | GTEx | DepMap | Descartes | 0.55 | 30.11 |
| STAT3 | 0.0012335 | 1656 | GTEx | DepMap | Descartes | 2.45 | 165.81 |
| EXTL2 | 0.0012229 | 1675 | GTEx | DepMap | Descartes | 0.48 | 45.41 |
| HS3ST3A1 | 0.0012119 | 1690 | GTEx | DepMap | Descartes | 0.06 | 4.76 |
| KDELR2 | 0.0010738 | 1917 | GTEx | DepMap | Descartes | 1.84 | 202.20 |
Descartes adrenocortical markers
Top 50 marker genes of adrenocortical cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/adrenocortical/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 3.13e-02
Mean rank of genes in gene set: 5152.47
Median rank of genes in gene set: 4124.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CLU | 0.0095567 | 5 | GTEx | DepMap | Descartes | 14.47 | 1772.86 |
| DNER | 0.0062353 | 31 | GTEx | DepMap | Descartes | 1.34 | 137.58 |
| GSTA4 | 0.0024232 | 627 | GTEx | DepMap | Descartes | 1.36 | 241.26 |
| HMGCS1 | 0.0022998 | 674 | GTEx | DepMap | Descartes | 0.92 | 42.90 |
| JAKMIP2 | 0.0020847 | 798 | GTEx | DepMap | Descartes | 0.73 | 23.99 |
| MSMO1 | 0.0017908 | 1020 | GTEx | DepMap | Descartes | 0.60 | 71.25 |
| FDPS | 0.0016702 | 1125 | GTEx | DepMap | Descartes | 2.01 | 270.63 |
| INHA | 0.0012291 | 1659 | GTEx | DepMap | Descartes | 0.08 | 17.23 |
| PEG3 | 0.0011722 | 1754 | GTEx | DepMap | Descartes | 0.42 | NA |
| FRMD5 | 0.0010426 | 1979 | GTEx | DepMap | Descartes | 0.11 | 6.37 |
| HMGCR | 0.0008824 | 2331 | GTEx | DepMap | Descartes | 0.34 | 21.88 |
| TM7SF2 | 0.0006727 | 2868 | GTEx | DepMap | Descartes | 0.38 | 54.29 |
| SGCZ | 0.0006227 | 3050 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SLC1A2 | 0.0005839 | 3200 | GTEx | DepMap | Descartes | 0.15 | 3.73 |
| PDE10A | 0.0005432 | 3349 | GTEx | DepMap | Descartes | 0.15 | 5.57 |
| IGF1R | 0.0004874 | 3551 | GTEx | DepMap | Descartes | 0.29 | 7.23 |
| POR | 0.0003769 | 4035 | GTEx | DepMap | Descartes | 0.43 | 51.86 |
| DHCR24 | 0.0003627 | 4100 | GTEx | DepMap | Descartes | 0.25 | 11.62 |
| SCAP | 0.0003533 | 4149 | GTEx | DepMap | Descartes | 0.21 | 15.51 |
| DHCR7 | 0.0003282 | 4270 | GTEx | DepMap | Descartes | 0.15 | 13.30 |
| CYB5B | 0.0002425 | 4704 | GTEx | DepMap | Descartes | 0.83 | 53.92 |
| SLC16A9 | 0.0000726 | 5785 | GTEx | DepMap | Descartes | 0.06 | 3.98 |
| SH3PXD2B | -0.0000347 | 6607 | GTEx | DepMap | Descartes | 0.09 | 4.39 |
| FREM2 | -0.0000645 | 6868 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| FDXR | -0.0001128 | 7390 | GTEx | DepMap | Descartes | 0.25 | 27.17 |
| SCARB1 | -0.0001684 | 8019 | GTEx | DepMap | Descartes | 0.16 | 9.31 |
| BAIAP2L1 | -0.0002061 | 8422 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| NPC1 | -0.0002734 | 9065 | GTEx | DepMap | Descartes | 0.09 | 6.28 |
| GRAMD1B | -0.0002867 | 9177 | GTEx | DepMap | Descartes | 0.04 | 1.26 |
| LDLR | -0.0003637 | 9808 | GTEx | DepMap | Descartes | 0.17 | 8.96 |
Descartes chromaffin markers
Top 50 marker genes of chromaffin cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/chromaffin/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.09e-16
Mean rank of genes in gene set: 1636.34
Median rank of genes in gene set: 622
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| NTRK1 | 0.0104639 | 2 | GTEx | DepMap | Descartes | 4.14 | 465.49 |
| TUBA1A | 0.0078693 | 13 | GTEx | DepMap | Descartes | 93.48 | 14182.12 |
| MLLT11 | 0.0077011 | 14 | GTEx | DepMap | Descartes | 27.14 | 3321.19 |
| STMN2 | 0.0067652 | 22 | GTEx | DepMap | Descartes | 70.70 | 10962.20 |
| TUBB2B | 0.0064973 | 29 | GTEx | DepMap | Descartes | 28.88 | 4404.20 |
| GAP43 | 0.0060681 | 35 | GTEx | DepMap | Descartes | 16.43 | 2557.78 |
| TUBB2A | 0.0060213 | 37 | GTEx | DepMap | Descartes | 19.74 | 3476.18 |
| MAP1B | 0.0058948 | 41 | GTEx | DepMap | Descartes | 24.19 | 598.30 |
| CNTFR | 0.0048895 | 97 | GTEx | DepMap | Descartes | 2.63 | 409.44 |
| REEP1 | 0.0047212 | 113 | GTEx | DepMap | Descartes | 1.74 | 130.90 |
| RGMB | 0.0044331 | 139 | GTEx | DepMap | Descartes | 2.01 | 137.62 |
| MARCH11 | 0.0040332 | 190 | GTEx | DepMap | Descartes | 0.99 | NA |
| TMEFF2 | 0.0035797 | 260 | GTEx | DepMap | Descartes | 1.88 | 171.88 |
| ISL1 | 0.0035492 | 266 | GTEx | DepMap | Descartes | 2.36 | 307.91 |
| BASP1 | 0.0034964 | 283 | GTEx | DepMap | Descartes | 15.23 | 2387.66 |
| RBFOX1 | 0.0030328 | 374 | GTEx | DepMap | Descartes | 0.78 | 50.63 |
| TMEM132C | 0.0025774 | 541 | GTEx | DepMap | Descartes | 0.21 | 13.85 |
| GREM1 | 0.0025502 | 553 | GTEx | DepMap | Descartes | 0.23 | 5.26 |
| SLC6A2 | 0.0025330 | 566 | GTEx | DepMap | Descartes | 0.43 | 44.35 |
| ANKFN1 | 0.0024603 | 606 | GTEx | DepMap | Descartes | 0.19 | 16.76 |
| HS3ST5 | 0.0024294 | 622 | GTEx | DepMap | Descartes | 0.61 | 48.72 |
| NPY | 0.0024235 | 626 | GTEx | DepMap | Descartes | 5.71 | 2161.52 |
| RPH3A | 0.0017485 | 1049 | GTEx | DepMap | Descartes | 0.13 | 9.49 |
| MAB21L1 | 0.0015512 | 1256 | GTEx | DepMap | Descartes | 1.93 | 194.98 |
| GAL | 0.0015416 | 1267 | GTEx | DepMap | Descartes | 5.74 | 2217.18 |
| MAB21L2 | 0.0014616 | 1362 | GTEx | DepMap | Descartes | 0.33 | 36.55 |
| IL7 | 0.0013960 | 1446 | GTEx | DepMap | Descartes | 1.89 | 264.21 |
| CCND1 | 0.0013064 | 1546 | GTEx | DepMap | Descartes | 7.46 | 536.83 |
| KCNB2 | 0.0011925 | 1723 | GTEx | DepMap | Descartes | 0.17 | 14.19 |
| FAT3 | 0.0011547 | 1775 | GTEx | DepMap | Descartes | 0.10 | 1.87 |
Descartes Vascular_endothelial markers
Top 50 marker genes of Vascular_endothelial cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/vascular_endothelial/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 8808.66
Median rank of genes in gene set: 9585.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| EFNB2 | 0.0006074 | 3102 | GTEx | DepMap | Descartes | 0.47 | 26.38 |
| EHD3 | 0.0005949 | 3144 | GTEx | DepMap | Descartes | 0.11 | 8.22 |
| NR5A2 | 0.0003658 | 4085 | GTEx | DepMap | Descartes | 0.03 | 2.07 |
| RAMP2 | 0.0002002 | 4968 | GTEx | DepMap | Descartes | 1.02 | 361.98 |
| HYAL2 | 0.0001206 | 5470 | GTEx | DepMap | Descartes | 0.34 | 24.22 |
| NOTCH4 | 0.0000202 | 6178 | GTEx | DepMap | Descartes | 0.22 | 9.70 |
| MYRIP | -0.0000207 | 6486 | GTEx | DepMap | Descartes | 0.02 | 1.01 |
| CYP26B1 | -0.0000593 | 6814 | GTEx | DepMap | Descartes | 0.01 | 0.54 |
| GALNT15 | -0.0001378 | 7675 | GTEx | DepMap | Descartes | 0.00 | NA |
| SHANK3 | -0.0001694 | 8030 | GTEx | DepMap | Descartes | 0.04 | 1.22 |
| ESM1 | -0.0001790 | 8139 | GTEx | DepMap | Descartes | 0.00 | 0.62 |
| CDH13 | -0.0002073 | 8431 | GTEx | DepMap | Descartes | 0.02 | 0.76 |
| CRHBP | -0.0002757 | 9088 | GTEx | DepMap | Descartes | 0.00 | 0.45 |
| TEK | -0.0002938 | 9242 | GTEx | DepMap | Descartes | 0.01 | 0.31 |
| SHE | -0.0003018 | 9304 | GTEx | DepMap | Descartes | 0.01 | 0.41 |
| PODXL | -0.0003021 | 9306 | GTEx | DepMap | Descartes | 0.05 | 2.93 |
| FLT4 | -0.0003131 | 9398 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| IRX3 | -0.0003177 | 9439 | GTEx | DepMap | Descartes | 0.02 | 1.88 |
| KDR | -0.0003337 | 9565 | GTEx | DepMap | Descartes | 0.01 | 0.50 |
| BTNL9 | -0.0003382 | 9606 | GTEx | DepMap | Descartes | 0.01 | 0.97 |
| CHRM3 | -0.0003634 | 9805 | GTEx | DepMap | Descartes | 0.11 | 2.16 |
| NPR1 | -0.0003665 | 9828 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ID1 | -0.0003734 | 9868 | GTEx | DepMap | Descartes | 0.12 | 31.80 |
| ROBO4 | -0.0003834 | 9944 | GTEx | DepMap | Descartes | 0.02 | 1.49 |
| F8 | -0.0003905 | 9999 | GTEx | DepMap | Descartes | 0.00 | 0.09 |
| RASIP1 | -0.0003924 | 10009 | GTEx | DepMap | Descartes | 0.01 | 0.36 |
| CEACAM1 | -0.0003952 | 10026 | GTEx | DepMap | Descartes | 0.02 | 3.25 |
| SLCO2A1 | -0.0003970 | 10039 | GTEx | DepMap | Descartes | 0.01 | 0.22 |
| MMRN2 | -0.0003994 | 10049 | GTEx | DepMap | Descartes | 0.00 | 0.18 |
| KANK3 | -0.0004316 | 10233 | GTEx | DepMap | Descartes | 0.00 | 0.17 |
Descartes stromal markers
Top 50 marker genes of stromal cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/stromal/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 9135.52
Median rank of genes in gene set: 10009
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| PCOLCE | 0.0013016 | 1557 | GTEx | DepMap | Descartes | 0.96 | 198.96 |
| PRICKLE1 | 0.0009289 | 2216 | GTEx | DepMap | Descartes | 0.22 | 12.15 |
| FREM1 | 0.0006849 | 2828 | GTEx | DepMap | Descartes | 0.03 | 1.18 |
| GAS2 | 0.0001257 | 5436 | GTEx | DepMap | Descartes | 0.01 | 0.71 |
| ADAMTS2 | 0.0000960 | 5632 | GTEx | DepMap | Descartes | 0.07 | 3.21 |
| ELN | 0.0000038 | 6294 | GTEx | DepMap | Descartes | 1.05 | 85.04 |
| GLI2 | -0.0000989 | 7232 | GTEx | DepMap | Descartes | 0.00 | 0.11 |
| CDH11 | -0.0001340 | 7631 | GTEx | DepMap | Descartes | 0.06 | 3.18 |
| LAMC3 | -0.0001541 | 7863 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ABCC9 | -0.0001889 | 8250 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ITGA11 | -0.0001952 | 8327 | GTEx | DepMap | Descartes | 0.01 | 0.25 |
| EDNRA | -0.0002158 | 8512 | GTEx | DepMap | Descartes | 0.00 | 0.56 |
| COL27A1 | -0.0002252 | 8609 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PCDH18 | -0.0002778 | 9113 | GTEx | DepMap | Descartes | 0.03 | 1.64 |
| ADAMTSL3 | -0.0002919 | 9224 | GTEx | DepMap | Descartes | 0.01 | 0.27 |
| PAMR1 | -0.0003250 | 9492 | GTEx | DepMap | Descartes | 0.02 | 1.96 |
| LRRC17 | -0.0003334 | 9561 | GTEx | DepMap | Descartes | 0.01 | 1.05 |
| SFRP2 | -0.0003494 | 9694 | GTEx | DepMap | Descartes | 0.00 | 0.75 |
| SCARA5 | -0.0003517 | 9708 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CLDN11 | -0.0003596 | 9773 | GTEx | DepMap | Descartes | 0.01 | 1.50 |
| DKK2 | -0.0003686 | 9845 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ABCA6 | -0.0003910 | 10004 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| C7 | -0.0003931 | 10014 | GTEx | DepMap | Descartes | 0.01 | 0.82 |
| ACTA2 | -0.0004179 | 10164 | GTEx | DepMap | Descartes | 0.05 | 10.28 |
| BICC1 | -0.0004239 | 10196 | GTEx | DepMap | Descartes | 0.03 | 1.72 |
| COL6A3 | -0.0004272 | 10214 | GTEx | DepMap | Descartes | 0.13 | 4.04 |
| RSPO3 | -0.0004285 | 10221 | GTEx | DepMap | Descartes | 0.00 | NA |
| IGFBP3 | -0.0004457 | 10309 | GTEx | DepMap | Descartes | 0.04 | 5.54 |
| HHIP | -0.0004590 | 10396 | GTEx | DepMap | Descartes | 0.02 | 0.37 |
| ISLR | -0.0004695 | 10463 | GTEx | DepMap | Descartes | 0.01 | 1.20 |
Descartes sympathoblasts markers
Top 50 marker genes of sympathoblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/sympathoblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 6.32e-05
Mean rank of genes in gene set: 4025.68
Median rank of genes in gene set: 2199.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CHGA | 0.0072050 | 16 | GTEx | DepMap | Descartes | 4.25 | 662.30 |
| C1QL1 | 0.0068111 | 21 | GTEx | DepMap | Descartes | 1.56 | 283.35 |
| PCSK1N | 0.0067364 | 23 | GTEx | DepMap | Descartes | 14.49 | 4392.91 |
| SLC18A1 | 0.0056771 | 48 | GTEx | DepMap | Descartes | 1.28 | 137.40 |
| CHGB | 0.0041548 | 174 | GTEx | DepMap | Descartes | 12.23 | 1517.14 |
| ARC | 0.0039038 | 206 | GTEx | DepMap | Descartes | 0.94 | 104.22 |
| PACRG | 0.0038926 | 209 | GTEx | DepMap | Descartes | 0.25 | 46.04 |
| MGAT4C | 0.0035369 | 270 | GTEx | DepMap | Descartes | 0.44 | 5.35 |
| GCH1 | 0.0021031 | 786 | GTEx | DepMap | Descartes | 0.65 | 76.36 |
| SLC24A2 | 0.0017990 | 1017 | GTEx | DepMap | Descartes | 0.04 | 1.11 |
| UNC80 | 0.0017552 | 1047 | GTEx | DepMap | Descartes | 0.39 | 8.94 |
| TBX20 | 0.0014465 | 1388 | GTEx | DepMap | Descartes | 0.06 | 9.71 |
| CDH12 | 0.0012911 | 1573 | GTEx | DepMap | Descartes | 0.13 | 10.19 |
| SLC35F3 | 0.0011866 | 1731 | GTEx | DepMap | Descartes | 0.08 | 8.23 |
| DGKK | 0.0011849 | 1735 | GTEx | DepMap | Descartes | 0.11 | 5.03 |
| AGBL4 | 0.0010814 | 1909 | GTEx | DepMap | Descartes | 0.05 | 5.14 |
| HTATSF1 | 0.0010061 | 2046 | GTEx | DepMap | Descartes | 0.75 | 73.27 |
| FAM155A | 0.0009895 | 2073 | GTEx | DepMap | Descartes | 0.27 | 9.05 |
| ROBO1 | 0.0009461 | 2169 | GTEx | DepMap | Descartes | 0.28 | 12.35 |
| SORCS3 | 0.0009228 | 2230 | GTEx | DepMap | Descartes | 0.02 | 0.29 |
| KCTD16 | 0.0007236 | 2728 | GTEx | DepMap | Descartes | 0.61 | 13.27 |
| CDH18 | 0.0006742 | 2858 | GTEx | DepMap | Descartes | 0.16 | 9.47 |
| PCSK2 | 0.0006620 | 2911 | GTEx | DepMap | Descartes | 0.09 | 5.21 |
| LAMA3 | 0.0002344 | 4750 | GTEx | DepMap | Descartes | 0.05 | 1.42 |
| EML6 | 0.0001873 | 5040 | GTEx | DepMap | Descartes | 0.07 | 2.11 |
| ST18 | 0.0000818 | 5731 | GTEx | DepMap | Descartes | 0.00 | 0.08 |
| SPOCK3 | -0.0000600 | 6828 | GTEx | DepMap | Descartes | 0.22 | 19.45 |
| CNTN3 | -0.0000731 | 6958 | GTEx | DepMap | Descartes | 0.01 | 0.18 |
| CCSER1 | -0.0001665 | 7997 | GTEx | DepMap | Descartes | 0.04 | NA |
| KSR2 | -0.0001678 | 8010 | GTEx | DepMap | Descartes | 0.01 | 0.06 |
Descartes erythroblasts markers
Top 50 marker genes of erythroblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/erythroblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 7.75e-01
Mean rank of genes in gene set: 6782.97
Median rank of genes in gene set: 8054
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| SOX6 | 0.0031936 | 334 | GTEx | DepMap | Descartes | 0.35 | 13.20 |
| TSPAN5 | 0.0023006 | 673 | GTEx | DepMap | Descartes | 0.81 | 63.62 |
| FECH | 0.0019802 | 870 | GTEx | DepMap | Descartes | 0.45 | 17.73 |
| SNCA | 0.0014463 | 1389 | GTEx | DepMap | Descartes | 0.95 | 82.57 |
| SPTB | 0.0011293 | 1832 | GTEx | DepMap | Descartes | 0.11 | 3.74 |
| TMCC2 | 0.0007653 | 2599 | GTEx | DepMap | Descartes | 0.14 | 9.77 |
| TRAK2 | 0.0006383 | 2986 | GTEx | DepMap | Descartes | 0.30 | 15.14 |
| XPO7 | 0.0005453 | 3342 | GTEx | DepMap | Descartes | 0.32 | 19.90 |
| ANK1 | 0.0004312 | 3807 | GTEx | DepMap | Descartes | 0.19 | 6.70 |
| SLC25A21 | 0.0003192 | 4316 | GTEx | DepMap | Descartes | 0.00 | 0.20 |
| RAPGEF2 | 0.0002633 | 4592 | GTEx | DepMap | Descartes | 0.22 | 8.05 |
| RHD | -0.0000518 | 6749 | GTEx | DepMap | Descartes | 0.00 | 0.32 |
| TFR2 | -0.0000968 | 7214 | GTEx | DepMap | Descartes | 0.14 | 12.04 |
| ABCB10 | -0.0001123 | 7382 | GTEx | DepMap | Descartes | 0.14 | 10.72 |
| CPOX | -0.0001714 | 8054 | GTEx | DepMap | Descartes | 0.06 | 5.96 |
| ALAS2 | -0.0001826 | 8178 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| RGS6 | -0.0001913 | 8276 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| GCLC | -0.0002523 | 8868 | GTEx | DepMap | Descartes | 0.08 | 5.66 |
| SLC4A1 | -0.0002564 | 8911 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| DENND4A | -0.0002735 | 9066 | GTEx | DepMap | Descartes | 0.20 | 7.46 |
| SLC25A37 | -0.0002776 | 9109 | GTEx | DepMap | Descartes | 0.53 | 34.74 |
| MARCH3 | -0.0003469 | 9674 | GTEx | DepMap | Descartes | 0.04 | NA |
| SELENBP1 | -0.0003559 | 9742 | GTEx | DepMap | Descartes | 0.01 | 0.50 |
| EPB41 | -0.0004040 | 10091 | GTEx | DepMap | Descartes | 0.35 | 17.12 |
| MICAL2 | -0.0005045 | 10657 | GTEx | DepMap | Descartes | 0.01 | 0.38 |
| SPECC1 | -0.0006413 | 11232 | GTEx | DepMap | Descartes | 0.00 | 0.08 |
| CAT | -0.0010211 | 12028 | GTEx | DepMap | Descartes | 0.16 | 21.16 |
| BLVRB | -0.0012670 | 12251 | GTEx | DepMap | Descartes | 0.02 | 3.86 |
| GYPC | -0.0019743 | 12484 | GTEx | DepMap | Descartes | 0.01 | 1.82 |
| NA | NA | NA | NA | NA | NA | NA | NA |
Descartes myeloid markers
Top 50 marker genes of myeloid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/myeloid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 10622.32
Median rank of genes in gene set: 11571.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CD163L1 | 0.0046711 | 118 | GTEx | DepMap | Descartes | 1.63 | 108.67 |
| FMN1 | 0.0010777 | 1914 | GTEx | DepMap | Descartes | 0.38 | 9.33 |
| HRH1 | 0.0002027 | 4953 | GTEx | DepMap | Descartes | 0.04 | 2.35 |
| WWP1 | -0.0001631 | 7960 | GTEx | DepMap | Descartes | 0.12 | 7.89 |
| RBPJ | -0.0003813 | 9925 | GTEx | DepMap | Descartes | 0.80 | 39.39 |
| SLC1A3 | -0.0003996 | 10051 | GTEx | DepMap | Descartes | 0.01 | 0.57 |
| ATP8B4 | -0.0004017 | 10071 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| RGL1 | -0.0004028 | 10081 | GTEx | DepMap | Descartes | 0.02 | 0.79 |
| MERTK | -0.0004113 | 10135 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SPP1 | -0.0004343 | 10249 | GTEx | DepMap | Descartes | 0.88 | 145.81 |
| SLCO2B1 | -0.0004360 | 10260 | GTEx | DepMap | Descartes | 0.01 | 0.17 |
| MSR1 | -0.0004375 | 10271 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CTSB | -0.0005139 | 10712 | GTEx | DepMap | Descartes | 1.73 | 138.90 |
| LGMN | -0.0005259 | 10764 | GTEx | DepMap | Descartes | 0.20 | 27.66 |
| ADAP2 | -0.0005690 | 10939 | GTEx | DepMap | Descartes | 0.01 | 0.83 |
| SLC9A9 | -0.0005966 | 11042 | GTEx | DepMap | Descartes | 0.00 | 0.37 |
| ABCA1 | -0.0006505 | 11260 | GTEx | DepMap | Descartes | 0.01 | 0.36 |
| SFMBT2 | -0.0006907 | 11376 | GTEx | DepMap | Descartes | 0.00 | 0.12 |
| MS4A4A | -0.0007570 | 11567 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| AXL | -0.0007614 | 11576 | GTEx | DepMap | Descartes | 0.00 | 0.09 |
| CD163 | -0.0008151 | 11714 | GTEx | DepMap | Descartes | 0.00 | 0.14 |
| FGD2 | -0.0008660 | 11789 | GTEx | DepMap | Descartes | 0.01 | 0.51 |
| ITPR2 | -0.0008885 | 11836 | GTEx | DepMap | Descartes | 0.02 | 0.29 |
| CSF1R | -0.0009192 | 11884 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CTSD | -0.0009214 | 11890 | GTEx | DepMap | Descartes | 0.50 | 75.74 |
| MARCH1 | -0.0009998 | 12005 | GTEx | DepMap | Descartes | 0.02 | NA |
| TGFBI | -0.0010290 | 12044 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CD14 | -0.0010778 | 12106 | GTEx | DepMap | Descartes | 0.03 | 5.59 |
| CPVL | -0.0011478 | 12158 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CYBB | -0.0011547 | 12164 | GTEx | DepMap | Descartes | 0.01 | 0.62 |
Descartes Schwann markers
Top 50 marker genes of Schwann cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/schwann/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.15e-01
Mean rank of genes in gene set: 7022.86
Median rank of genes in gene set: 8235.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| PMP22 | 0.0103218 | 4 | GTEx | DepMap | Descartes | 8.99 | 1573.97 |
| GFRA3 | 0.0062613 | 30 | GTEx | DepMap | Descartes | 2.98 | 458.95 |
| SCN7A | 0.0047874 | 109 | GTEx | DepMap | Descartes | 1.00 | 46.31 |
| LRRTM4 | 0.0046504 | 119 | GTEx | DepMap | Descartes | 0.59 | 53.86 |
| DST | 0.0043316 | 153 | GTEx | DepMap | Descartes | 3.37 | 50.26 |
| SFRP1 | 0.0040183 | 192 | GTEx | DepMap | Descartes | 2.33 | 162.18 |
| MARCKS | 0.0035595 | 265 | GTEx | DepMap | Descartes | 9.50 | 666.01 |
| EGFLAM | 0.0019845 | 863 | GTEx | DepMap | Descartes | 0.13 | 7.57 |
| LAMA4 | 0.0016374 | 1159 | GTEx | DepMap | Descartes | 0.40 | 18.52 |
| SORCS1 | 0.0016301 | 1172 | GTEx | DepMap | Descartes | 0.15 | 5.74 |
| PPP2R2B | 0.0016171 | 1191 | GTEx | DepMap | Descartes | 0.53 | 14.61 |
| FIGN | 0.0004792 | 3586 | GTEx | DepMap | Descartes | 0.10 | 3.07 |
| TRPM3 | 0.0002504 | 4665 | GTEx | DepMap | Descartes | 0.10 | 2.48 |
| NRXN1 | 0.0001462 | 5281 | GTEx | DepMap | Descartes | 0.69 | 21.32 |
| GRIK3 | 0.0001258 | 5435 | GTEx | DepMap | Descartes | 0.01 | 0.84 |
| ERBB3 | 0.0001155 | 5503 | GTEx | DepMap | Descartes | 0.02 | 0.85 |
| MPZ | 0.0000382 | 6041 | GTEx | DepMap | Descartes | 0.03 | 5.46 |
| NRXN3 | -0.0000766 | 6994 | GTEx | DepMap | Descartes | 0.03 | 0.80 |
| PLCE1 | -0.0001405 | 7711 | GTEx | DepMap | Descartes | 0.07 | 1.86 |
| LAMB1 | -0.0001509 | 7819 | GTEx | DepMap | Descartes | 0.20 | 9.25 |
| PTPRZ1 | -0.0001748 | 8088 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| HMGA2 | -0.0001863 | 8219 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| COL25A1 | -0.0001890 | 8252 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| XKR4 | -0.0001945 | 8313 | GTEx | DepMap | Descartes | 0.08 | 1.21 |
| PLP1 | -0.0002931 | 9234 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| STARD13 | -0.0003020 | 9305 | GTEx | DepMap | Descartes | 0.01 | 0.35 |
| IL1RAPL1 | -0.0003033 | 9319 | GTEx | DepMap | Descartes | 0.01 | 0.25 |
| IL1RAPL2 | -0.0003925 | 10010 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ADAMTS5 | -0.0004276 | 10217 | GTEx | DepMap | Descartes | 0.00 | 0.07 |
| PTN | -0.0004416 | 10292 | GTEx | DepMap | Descartes | 2.37 | 434.59 |
Descartes Megakaryocytes markers
Top 50 marker genes of Megakaryocytes cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/megakaryocytes/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 9053.76
Median rank of genes in gene set: 10474
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| PRKAR2B | 0.0041268 | 179 | GTEx | DepMap | Descartes | 1.36 | 109.85 |
| CD9 | 0.0028485 | 439 | GTEx | DepMap | Descartes | 2.95 | 528.72 |
| RAB27B | 0.0009241 | 2228 | GTEx | DepMap | Descartes | 0.19 | 8.47 |
| TUBB1 | 0.0008492 | 2400 | GTEx | DepMap | Descartes | 0.04 | 3.43 |
| PDE3A | 0.0007802 | 2559 | GTEx | DepMap | Descartes | 0.07 | 2.16 |
| STON2 | 0.0005987 | 3129 | GTEx | DepMap | Descartes | 0.10 | 6.45 |
| ACTN1 | 0.0004548 | 3697 | GTEx | DepMap | Descartes | 0.53 | 35.90 |
| ZYX | 0.0003103 | 4352 | GTEx | DepMap | Descartes | 0.78 | 111.83 |
| ACTB | 0.0000936 | 5649 | GTEx | DepMap | Descartes | 57.09 | 7613.36 |
| ITGB3 | 0.0000432 | 6007 | GTEx | DepMap | Descartes | 0.01 | 0.34 |
| ITGA2B | -0.0000148 | 6441 | GTEx | DepMap | Descartes | 0.04 | 3.69 |
| VCL | -0.0000752 | 6979 | GTEx | DepMap | Descartes | 0.16 | 6.87 |
| INPP4B | -0.0001581 | 7903 | GTEx | DepMap | Descartes | 0.09 | 4.01 |
| ANGPT1 | -0.0002123 | 8477 | GTEx | DepMap | Descartes | 0.00 | 0.15 |
| TRPC6 | -0.0002231 | 8587 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| GP1BA | -0.0002366 | 8708 | GTEx | DepMap | Descartes | 0.01 | 1.45 |
| SLC24A3 | -0.0002451 | 8794 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ARHGAP6 | -0.0003036 | 9323 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MYLK | -0.0003899 | 9993 | GTEx | DepMap | Descartes | 0.01 | 0.44 |
| MMRN1 | -0.0004027 | 10079 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| LTBP1 | -0.0004217 | 10187 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MED12L | -0.0004695 | 10462 | GTEx | DepMap | Descartes | 0.01 | 0.19 |
| MCTP1 | -0.0004717 | 10474 | GTEx | DepMap | Descartes | 0.03 | 1.96 |
| UBASH3B | -0.0004813 | 10531 | GTEx | DepMap | Descartes | 0.04 | 1.67 |
| LIMS1 | -0.0005392 | 10826 | GTEx | DepMap | Descartes | 0.76 | 51.14 |
| P2RX1 | -0.0005403 | 10831 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| GSN | -0.0005731 | 10960 | GTEx | DepMap | Descartes | 0.28 | 14.13 |
| HIPK2 | -0.0005900 | 11014 | GTEx | DepMap | Descartes | 0.37 | 6.94 |
| PSTPIP2 | -0.0006694 | 11308 | GTEx | DepMap | Descartes | 0.00 | 0.15 |
| STOM | -0.0007992 | 11673 | GTEx | DepMap | Descartes | 0.13 | 13.71 |
Descartes Lyphoid markers
Top 50 marker genes of Lyphoid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/lymphoid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 10167.83
Median rank of genes in gene set: 12019
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TMSB10 | 0.0026151 | 525 | GTEx | DepMap | Descartes | 101.43 | 64770.97 |
| STK39 | 0.0024962 | 586 | GTEx | DepMap | Descartes | 0.77 | 66.95 |
| RAP1GAP2 | 0.0018096 | 1011 | GTEx | DepMap | Descartes | 0.48 | 24.13 |
| NCALD | 0.0010384 | 1985 | GTEx | DepMap | Descartes | 0.59 | 44.10 |
| GNG2 | 0.0005831 | 3202 | GTEx | DepMap | Descartes | 1.58 | 133.45 |
| FYN | 0.0004391 | 3765 | GTEx | DepMap | Descartes | 2.32 | 191.63 |
| CD44 | 0.0002893 | 4467 | GTEx | DepMap | Descartes | 2.31 | 133.55 |
| BCL2 | 0.0000132 | 6223 | GTEx | DepMap | Descartes | 0.39 | 15.96 |
| SCML4 | -0.0001574 | 7893 | GTEx | DepMap | Descartes | 0.31 | 21.20 |
| FOXP1 | -0.0003879 | 9975 | GTEx | DepMap | Descartes | 1.40 | 47.34 |
| SORL1 | -0.0005684 | 10937 | GTEx | DepMap | Descartes | 0.35 | 10.27 |
| DOCK10 | -0.0006759 | 11327 | GTEx | DepMap | Descartes | 0.13 | 5.79 |
| ITPKB | -0.0007028 | 11411 | GTEx | DepMap | Descartes | 0.00 | 0.06 |
| PITPNC1 | -0.0007923 | 11657 | GTEx | DepMap | Descartes | 0.14 | 6.24 |
| TOX | -0.0008041 | 11681 | GTEx | DepMap | Descartes | 0.13 | 7.98 |
| BACH2 | -0.0008329 | 11743 | GTEx | DepMap | Descartes | 0.11 | 3.75 |
| SAMD3 | -0.0008627 | 11783 | GTEx | DepMap | Descartes | 0.01 | 0.38 |
| ABLIM1 | -0.0009021 | 11856 | GTEx | DepMap | Descartes | 0.14 | 4.87 |
| LEF1 | -0.0009477 | 11932 | GTEx | DepMap | Descartes | 0.00 | 0.16 |
| SKAP1 | -0.0010084 | 12013 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PDE3B | -0.0010107 | 12016 | GTEx | DepMap | Descartes | 0.06 | 2.60 |
| MCTP2 | -0.0010174 | 12022 | GTEx | DepMap | Descartes | 0.00 | 0.05 |
| PLEKHA2 | -0.0010656 | 12088 | GTEx | DepMap | Descartes | 0.03 | 1.91 |
| ANKRD44 | -0.0011009 | 12123 | GTEx | DepMap | Descartes | 0.19 | 8.37 |
| CCL5 | -0.0011738 | 12178 | GTEx | DepMap | Descartes | 0.04 | 9.40 |
| PRKCH | -0.0012357 | 12221 | GTEx | DepMap | Descartes | 0.01 | 0.27 |
| ARID5B | -0.0012900 | 12264 | GTEx | DepMap | Descartes | 0.10 | 4.40 |
| CELF2 | -0.0013227 | 12282 | GTEx | DepMap | Descartes | 0.72 | 26.16 |
| IKZF1 | -0.0013479 | 12294 | GTEx | DepMap | Descartes | 0.01 | 0.26 |
| ARHGAP15 | -0.0013483 | 12295 | GTEx | DepMap | Descartes | 0.01 | 0.24 |
Below shows the ranks on this GEP for immune marker gene lists curated by CellTypist (https://www.celltypist.org/encyclopedia).
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Erythroid: Mid erythroid (curated markers)
middle erythroid cells in fetal liver:
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.12e-02
Mean rank of genes in gene set: 427
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| PRDX2 | 0.0038884 | 210 | GTEx | DepMap | Descartes | 6.53 | 1141.06 |
| KCNH2 | 0.0023708 | 644 | GTEx | DepMap | Descartes | 0.69 | 48.48 |
T cells: Tem/Effector helper T cells PD1+ (model markers)
CD4+ helper T lymphocyte subpopulation in the thymus which features the expression of programmed cell death protein 1 (PD-1):
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.67e-02
Mean rank of genes in gene set: 826
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| PANX2 | 0.0037409 | 226 | GTEx | DepMap | Descartes | 0.50 | 56.42 |
| INPP5J | 0.0014116 | 1426 | GTEx | DepMap | Descartes | 0.12 | 11.29 |
B cells: Transitional B cells (model markers)
immature B cell precursors in the bone marrow which connect Pre-B cells with mature naive B cells and are subject to the process of B cell selection:
Wilcoxon ranksum test P-value for gene set overrepresentation: 4.27e-02
Mean rank of genes in gene set: 44
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| GNG3 | 0.0057544 | 44 | GTEx | DepMap | Descartes | 4.39 | 1563.11 |
No detectable expression in this dataset: OR2A25