Program: 13. Erythroblasts.
Submit a comment on this gene expression program’s interpretation: CLICK
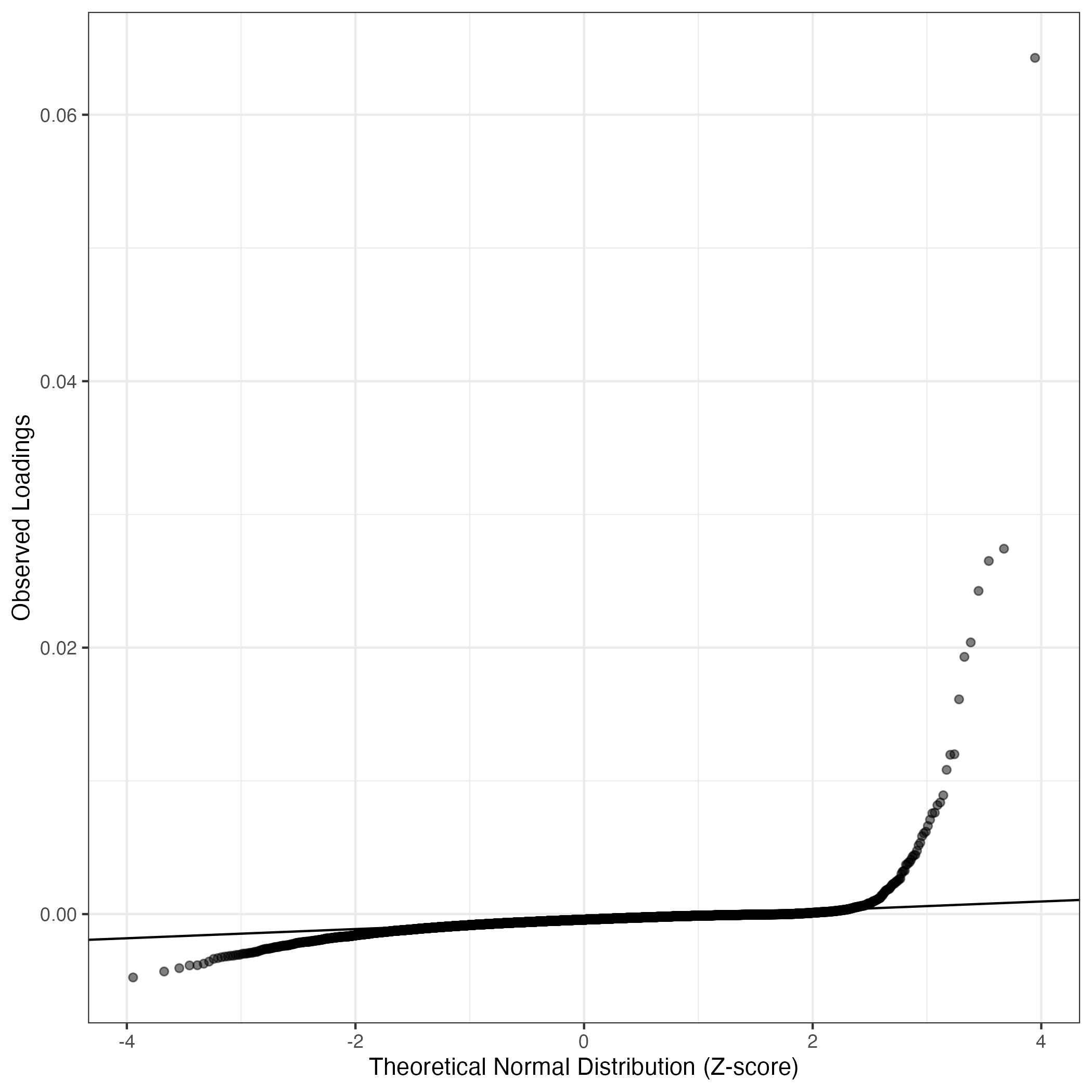
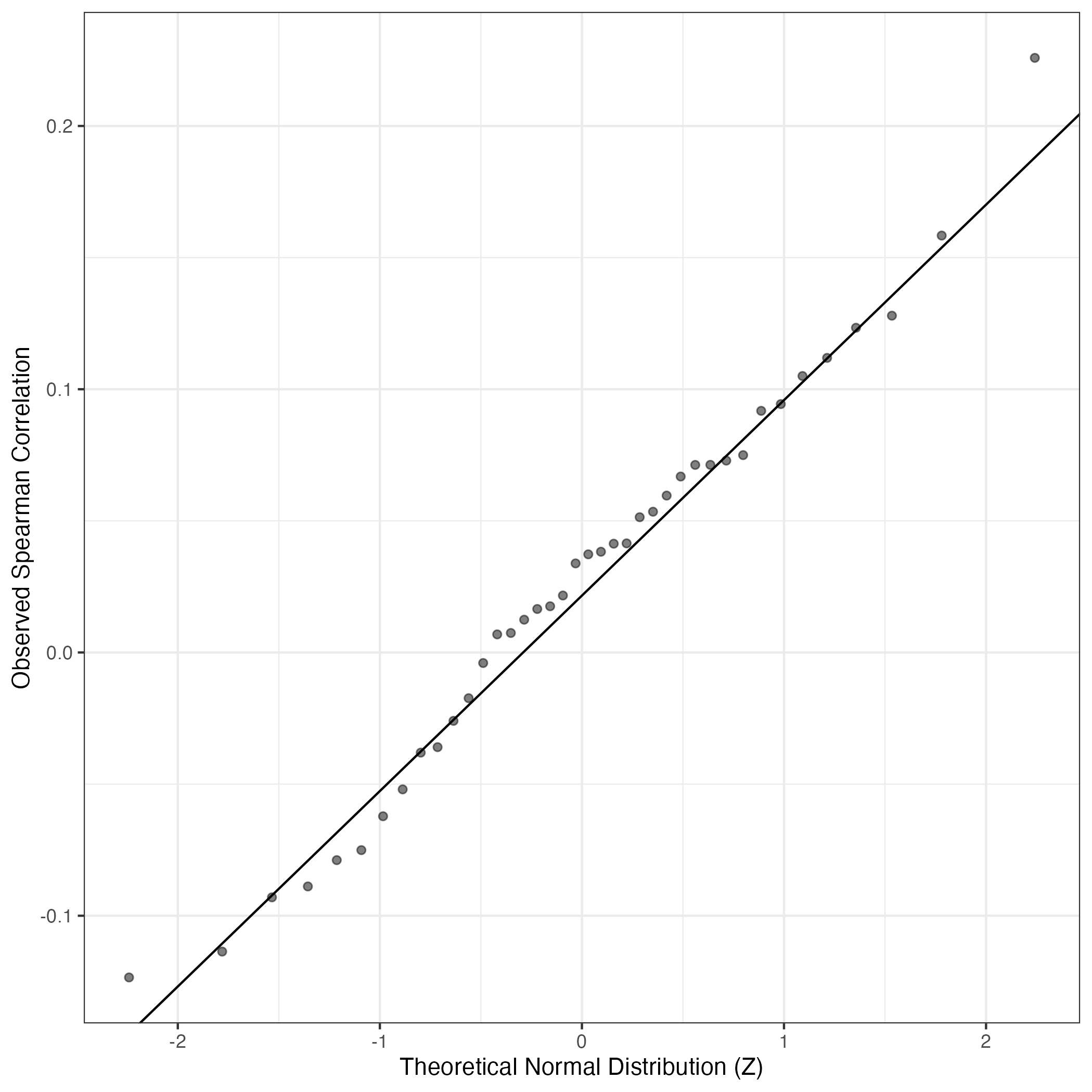
QQ-plot of gene loadings, averaged over both independent splits of the data
This plot highlights the relative contribution of each gene to the GEP
Top genes driving this program.
Note: Decartes website is buggy, try refreshing. Also, Decartes fetal adrenal data have been collected at specific time points (89-122 days), all possible cell types of interest may not be represented, do not overinterpret.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
| Gene | Loading | Gene.Name | GTEx | DepMap | Descartes | Mean.Counts | Mean.Tpm | |
|---|---|---|---|---|---|---|---|---|
| 1 | ALAS2 | 0.0642696 | 5’-aminolevulinate synthase 2 | GTEx | DepMap | Descartes | 10.13 | 555.57 |
| 2 | SELENBP1 | 0.0274233 | selenium binding protein 1 | GTEx | DepMap | Descartes | 1.11 | 34.19 |
| 3 | SLC25A37 | 0.0265062 | solute carrier family 25 member 37 | GTEx | DepMap | Descartes | 6.62 | 151.57 |
| 4 | KLC3 | 0.0242573 | kinesin light chain 3 | GTEx | DepMap | Descartes | 0.11 | 4.68 |
| 5 | SLC4A1 | 0.0203923 | solute carrier family 4 member 1 (Diego blood group) | GTEx | DepMap | Descartes | 0.57 | 10.08 |
| 6 | GMPR | 0.0193077 | guanosine monophosphate reductase | GTEx | DepMap | Descartes | 1.64 | 117.43 |
| 7 | NFE2 | 0.0161206 | nuclear factor, erythroid 2 | GTEx | DepMap | Descartes | 0.17 | 7.77 |
| 8 | MYL4 | 0.0119982 | myosin light chain 4 | GTEx | DepMap | Descartes | 0.53 | 60.55 |
| 9 | SNCA | 0.0119594 | synuclein alpha | GTEx | DepMap | Descartes | 4.38 | 137.78 |
| 10 | STRADB | 0.0108294 | STE20 related adaptor beta | GTEx | DepMap | Descartes | 1.72 | 76.15 |
| 11 | TRIM58 | 0.0089176 | tripartite motif containing 58 | GTEx | DepMap | Descartes | 0.32 | 5.83 |
| 12 | ARG1 | 0.0083776 | arginase 1 | GTEx | DepMap | Descartes | 0.04 | 2.44 |
| 13 | RHD | 0.0081725 | Rh blood group D antigen | GTEx | DepMap | Descartes | 0.09 | 3.24 |
| 14 | SLC14A1 | 0.0076134 | solute carrier family 14 member 1 (Kidd blood group) | GTEx | DepMap | Descartes | 0.21 | 4.48 |
| 15 | BPGM | 0.0075670 | bisphosphoglycerate mutase | GTEx | DepMap | Descartes | 1.53 | 69.30 |
| 16 | SLC25A39 | 0.0070849 | solute carrier family 25 member 39 | GTEx | DepMap | Descartes | 4.38 | 215.33 |
| 17 | GYPC | 0.0066174 | glycophorin C (Gerbich blood group) | GTEx | DepMap | Descartes | 4.53 | 296.75 |
| 18 | SMOX | 0.0061870 | spermine oxidase | GTEx | DepMap | Descartes | 0.49 | 16.25 |
| 19 | BLVRB | 0.0060679 | biliverdin reductase B | GTEx | DepMap | Descartes | 2.87 | 205.16 |
| 20 | UBB | 0.0058407 | ubiquitin B | GTEx | DepMap | Descartes | 22.26 | 2314.27 |
| 21 | ADIPOR1 | 0.0053585 | adiponectin receptor 1 | GTEx | DepMap | Descartes | 2.30 | 127.50 |
| 22 | MPP1 | 0.0051541 | MAGUK p55 scaffold protein 1 | GTEx | DepMap | Descartes | 1.04 | 40.99 |
| 23 | RAP1GAP | 0.0047549 | RAP1 GTPase activating protein | GTEx | DepMap | Descartes | 0.26 | 5.48 |
| 24 | GFI1B | 0.0044495 | growth factor independent 1B transcriptional repressor | GTEx | DepMap | Descartes | 0.02 | 0.60 |
| 25 | YBX3 | 0.0044226 | Y-box binding protein 3 | GTEx | DepMap | Descartes | 2.04 | NA |
| 26 | NCOA4 | 0.0043054 | nuclear receptor coactivator 4 | GTEx | DepMap | Descartes | 1.85 | 51.45 |
| 27 | NFIX | 0.0040876 | nuclear factor I X | GTEx | DepMap | Descartes | 0.17 | 2.59 |
| 28 | BNIP3L | 0.0038987 | BCL2 interacting protein 3 like | GTEx | DepMap | Descartes | 2.77 | 83.36 |
| 29 | FKBP8 | 0.0038749 | FKBP prolyl isomerase 8 | GTEx | DepMap | Descartes | 4.53 | 257.63 |
| 30 | DCAF12 | 0.0037220 | DDB1 and CUL4 associated factor 12 | GTEx | DepMap | Descartes | 0.70 | 20.10 |
| 31 | FAM178B | 0.0037072 | family with sequence similarity 178 member B | GTEx | DepMap | Descartes | 0.77 | 15.39 |
| 32 | FAM210B | 0.0032533 | family with sequence similarity 210 member B | GTEx | DepMap | Descartes | 1.55 | 61.02 |
| 33 | MAP2K3 | 0.0032470 | mitogen-activated protein kinase kinase 3 | GTEx | DepMap | Descartes | 0.70 | 31.78 |
| 34 | SMIM1 | 0.0031779 | small integral membrane protein 1 (Vel blood group) | GTEx | DepMap | Descartes | 0.62 | NA |
| 35 | FBXO7 | 0.0030217 | F-box protein 7 | GTEx | DepMap | Descartes | 1.72 | 23.66 |
| 36 | SMIM5 | 0.0026517 | small integral membrane protein 5 | GTEx | DepMap | Descartes | 0.04 | NA |
| 37 | DMTN | 0.0026395 | dematin actin binding protein | GTEx | DepMap | Descartes | 0.87 | NA |
| 38 | PLEK2 | 0.0025554 | pleckstrin 2 | GTEx | DepMap | Descartes | 0.09 | 6.39 |
| 39 | TMPRSS9 | 0.0024638 | transmembrane serine protease 9 | GTEx | DepMap | Descartes | 0.02 | 0.35 |
| 40 | CCNDBP1 | 0.0024631 | cyclin D1 binding protein 1 | GTEx | DepMap | Descartes | 1.26 | 38.83 |
| 41 | R3HDM4 | 0.0023779 | R3H domain containing 4 | GTEx | DepMap | Descartes | 0.96 | 66.45 |
| 42 | RNF10 | 0.0022874 | ring finger protein 10 | GTEx | DepMap | Descartes | 1.43 | 41.29 |
| 43 | MKRN1 | 0.0022791 | makorin ring finger protein 1 | GTEx | DepMap | Descartes | 1.66 | 49.13 |
| 44 | TUBB1 | 0.0022514 | tubulin beta 1 class VI | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| 45 | RSPO1 | 0.0021585 | R-spondin 1 | GTEx | DepMap | Descartes | 0.02 | 0.63 |
| 46 | PITHD1 | 0.0020609 | PITH domain containing 1 | GTEx | DepMap | Descartes | 0.96 | 51.64 |
| 47 | BCL2L1 | 0.0019883 | BCL2 like 1 | GTEx | DepMap | Descartes | 0.64 | 25.40 |
| 48 | PRODH | 0.0018877 | proline dehydrogenase 1 | GTEx | DepMap | Descartes | 0.02 | 0.40 |
| 49 | HPS1 | 0.0018803 | HPS1 biogenesis of lysosomal organelles complex 3 subunit 1 | GTEx | DepMap | Descartes | 0.43 | 11.69 |
| 50 | GUK1 | 0.0018549 | guanylate kinase 1 | GTEx | DepMap | Descartes | 4.64 | 204.67 |
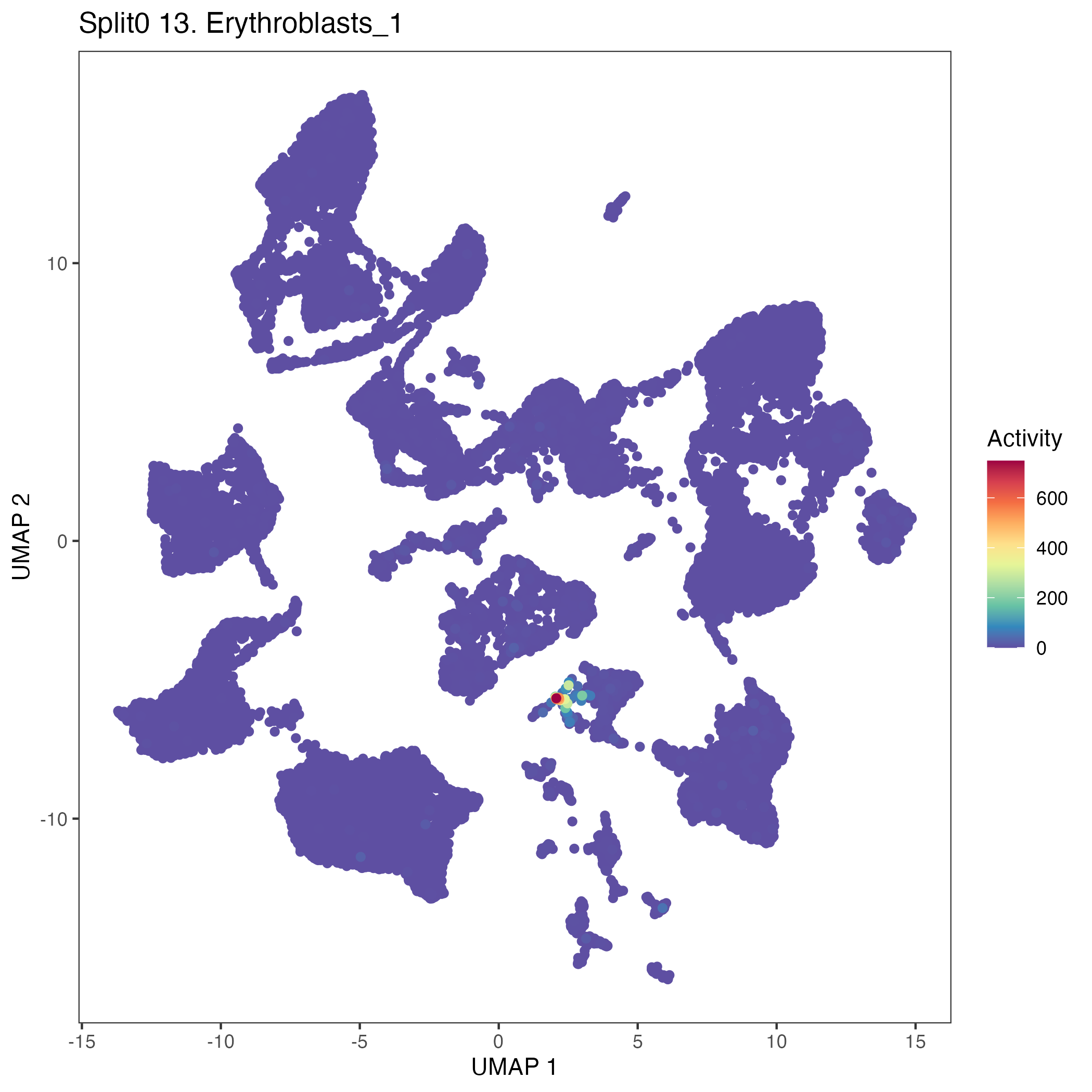
UMAP plots showing activity of gene expression program identified in GEP 13. Erythroblasts:
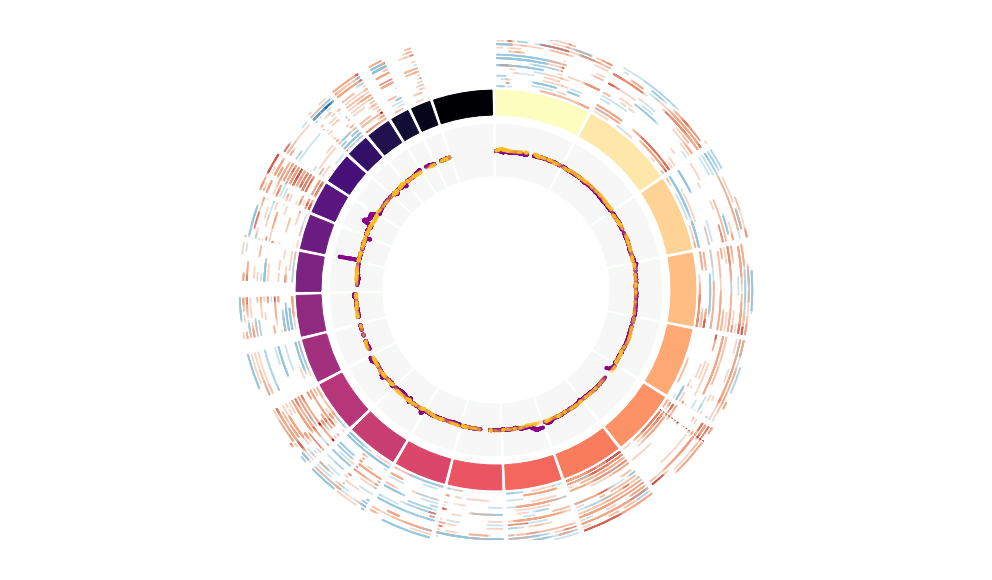
CNV Data procured from inferCNV.
Outer tracks are putative CNV regions (gains = red, losses = blue) for each patient
Inner track is expression data representing:
The top cells expressing this GEP (purple)
Random cells (n =50) from the reference set used in inferCNV (orange)
Gene set Enrichments for this program, caculated from top 50 genes
mSigDB Cell Types Gene Set:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| HU_FETAL_RETINA_BLOOD | 1.58e-39 | 81.22 | 43.79 | 1.06e-36 | 1.06e-36 | 31ALAS2, SELENBP1, SLC25A37, SLC4A1, GMPR, NFE2, MYL4, SNCA, STRADB, BPGM, SLC25A39, GYPC, BLVRB, UBB, ADIPOR1, MPP1, RAP1GAP, GFI1B, YBX3, NCOA4, DCAF12, FAM210B, SMIM1, FBXO7, SMIM5, DMTN, CCNDBP1, TUBB1, BCL2L1, HPS1, GUK1 |
282 |
| DESCARTES_FETAL_INTESTINE_ERYTHROBLASTS | 2.35e-29 | 71.01 | 37.77 | 7.90e-27 | 1.58e-26 | 22ALAS2, SELENBP1, SLC25A37, SLC4A1, GMPR, NFE2, MYL4, SNCA, STRADB, TRIM58, RHD, SLC14A1, BPGM, SLC25A39, BLVRB, GFI1B, DCAF12, FAM210B, SMIM1, DMTN, TUBB1, BCL2L1 |
160 |
| DESCARTES_FETAL_KIDNEY_ERYTHROBLASTS | 3.59e-27 | 62.35 | 32.96 | 8.03e-25 | 2.41e-24 | 21ALAS2, SELENBP1, SLC25A37, KLC3, SLC4A1, NFE2, MYL4, SNCA, TRIM58, ARG1, RHD, SLC14A1, BPGM, SLC25A39, BLVRB, GFI1B, FKBP8, DCAF12, MAP2K3, SMIM1, SMIM5 |
166 |
| DESCARTES_FETAL_PANCREAS_ERYTHROBLASTS | 1.29e-23 | 59.95 | 30.77 | 2.17e-21 | 8.68e-21 | 18ALAS2, SLC25A37, SLC4A1, GMPR, NFE2, MYL4, SNCA, TRIM58, RHD, SLC14A1, BPGM, SLC25A39, BLVRB, GFI1B, DCAF12, MAP2K3, SMIM1, PLEK2 |
135 |
| DESCARTES_FETAL_STOMACH_ERYTHROBLASTS | 1.77e-20 | 62.89 | 30.75 | 2.38e-18 | 1.19e-17 | 15ALAS2, SLC25A37, SLC4A1, NFE2, MYL4, SNCA, STRADB, TRIM58, RHD, SLC14A1, SLC25A39, BLVRB, DCAF12, MAP2K3, TUBB1 |
100 |
| DESCARTES_MAIN_FETAL_ERYTHROBLASTS | 5.39e-19 | 31.39 | 16.34 | 6.03e-17 | 3.62e-16 | 18ALAS2, SELENBP1, SLC25A37, SLC4A1, GMPR, SNCA, TRIM58, RHD, SLC14A1, SLC25A39, GYPC, BLVRB, BNIP3L, DCAF12, FAM178B, MAP2K3, FBXO7, DMTN |
242 |
| DESCARTES_FETAL_ADRENAL_ERYTHROBLASTS | 1.78e-11 | 30.02 | 13.03 | 1.49e-09 | 1.19e-08 | 10ALAS2, SELENBP1, SLC25A37, SLC4A1, ARG1, RHD, SLC14A1, SLC25A39, BLVRB, FAM178B |
114 |
| DESCARTES_FETAL_MUSCLE_ERYTHROBLASTS | 1.78e-09 | 22.48 | 9.40 | 1.19e-07 | 1.19e-06 | 9ALAS2, SELENBP1, SLC25A37, SLC4A1, RHD, SLC14A1, BLVRB, FAM210B, DMTN |
131 |
| DESCARTES_FETAL_LIVER_ERYTHROBLASTS | 1.19e-05 | 19.54 | 5.88 | 5.31e-04 | 7.96e-03 | 5ALAS2, SLC25A37, SLC4A1, RHD, SLC14A1 |
76 |
| TRAVAGLINI_LUNG_PLATELET_MEGAKARYOCYTE_CELL | 3.44e-11 | 10.98 | 5.70 | 2.56e-09 | 2.31e-08 | 17GMPR, NFE2, SNCA, TRIM58, SMOX, ADIPOR1, MPP1, GFI1B, NCOA4, FKBP8, MAP2K3, SMIM5, DMTN, R3HDM4, MKRN1, TUBB1, BCL2L1 |
604 |
| DESCARTES_FETAL_LUNG_MEGAKARYOCYTES | 3.90e-07 | 14.09 | 5.62 | 2.38e-05 | 2.62e-04 | 8GMPR, NFE2, TRIM58, RHD, SMOX, MPP1, GFI1B, TUBB1 |
177 |
| HAY_BONE_MARROW_ERYTHROBLAST | 1.26e-13 | 10.06 | 5.53 | 1.20e-11 | 8.43e-11 | 25ALAS2, SELENBP1, SLC25A37, SLC4A1, MYL4, SNCA, STRADB, RHD, SLC25A39, GYPC, BLVRB, MPP1, GFI1B, YBX3, NFIX, FAM178B, FAM210B, MAP2K3, SMIM1, FBXO7, DMTN, PLEK2, TMPRSS9, PITHD1, HPS1 |
1269 |
| DESCARTES_FETAL_HEART_MEGAKARYOCYTES | 4.85e-06 | 16.07 | 5.48 | 2.32e-04 | 3.25e-03 | 6NFE2, TRIM58, RHD, MPP1, GFI1B, TUBB1 |
112 |
| HAY_BONE_MARROW_PLATELET | 8.93e-07 | 10.43 | 4.41 | 4.99e-05 | 5.99e-04 | 9GMPR, NFE2, TRIM58, SMOX, NCOA4, SMIM5, R3HDM4, TUBB1, BCL2L1 |
272 |
| DESCARTES_FETAL_CEREBRUM_MEGAKARYOCYTES | 4.06e-06 | 7.53 | 3.34 | 2.10e-04 | 2.73e-03 | 10GMPR, MYL4, STRADB, TRIM58, RHD, GFI1B, DCAF12, SMIM5, DMTN, TUBB1 |
425 |
| DESCARTES_FETAL_SPLEEN_ERYTHROBLASTS | 2.25e-03 | 12.65 | 2.46 | 9.43e-02 | 1.00e+00 | 3SLC4A1, SLC14A1, FAM178B |
66 |
| ZHENG_CORD_BLOOD_C2_PUTATIVE_BASOPHIL_EOSINOPHIL_MAST_CELL_PROGENITOR | 6.42e-03 | 8.57 | 1.68 | 2.39e-01 | 1.00e+00 | 3FAM178B, SMIM1, FBXO7 |
96 |
| ZHENG_CORD_BLOOD_C1_PUTATIVE_MEGAKARYOCYTE_PROGENITOR | 6.79e-03 | 8.40 | 1.64 | 2.40e-01 | 1.00e+00 | 3GFI1B, DMTN, TUBB1 |
98 |
| ZHENG_CORD_BLOOD_C3_MEGAKARYOCYTE_ERYTHROID_PROGENITOR | 7.18e-03 | 8.22 | 1.61 | 2.41e-01 | 1.00e+00 | 3BLVRB, FAM178B, FBXO7 |
100 |
| TRAVAGLINI_LUNG_CAPILLARY_AEROCYTE_CELL | 1.20e-02 | 13.01 | 1.48 | 3.67e-01 | 1.00e+00 | 2SLC14A1, UBB |
42 |
Dowload full table
mSigDB Hallmark Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| HALLMARK_HEME_METABOLISM | 3.75e-27 | 55.14 | 29.44 | 1.87e-25 | 1.87e-25 | 22ALAS2, SELENBP1, SLC25A37, SLC4A1, NFE2, MYL4, SNCA, TRIM58, RHD, BPGM, GYPC, SMOX, BLVRB, ADIPOR1, MPP1, RAP1GAP, NCOA4, BNIP3L, MAP2K3, FBXO7, DMTN, MKRN1 |
200 |
| HALLMARK_XENOBIOTIC_METABOLISM | 7.71e-03 | 5.55 | 1.44 | 1.28e-01 | 3.85e-01 | 4ARG1, SMOX, BLVRB, RAP1GAP |
200 |
| HALLMARK_KRAS_SIGNALING_UP | 7.71e-03 | 5.55 | 1.44 | 1.28e-01 | 3.85e-01 | 4ARG1, BPGM, GYPC, PLEK2 |
200 |
| HALLMARK_APOPTOSIS | 1.32e-01 | 3.28 | 0.38 | 1.00e+00 | 1.00e+00 | 2BNIP3L, BCL2L1 |
161 |
| HALLMARK_HYPOXIA | 1.85e-01 | 2.63 | 0.31 | 1.00e+00 | 1.00e+00 | 2SELENBP1, BNIP3L |
200 |
| HALLMARK_INTERFERON_ALPHA_RESPONSE | 3.20e-01 | 2.66 | 0.07 | 1.00e+00 | 1.00e+00 | 1GMPR |
97 |
| HALLMARK_ANDROGEN_RESPONSE | 3.28e-01 | 2.58 | 0.06 | 1.00e+00 | 1.00e+00 | 1NCOA4 |
100 |
| HALLMARK_PI3K_AKT_MTOR_SIGNALING | 3.41e-01 | 2.45 | 0.06 | 1.00e+00 | 1.00e+00 | 1MAP2K3 |
105 |
| HALLMARK_DNA_REPAIR | 4.49e-01 | 1.71 | 0.04 | 1.00e+00 | 1.00e+00 | 1GUK1 |
150 |
| HALLMARK_IL2_STAT5_SIGNALING | 5.45e-01 | 1.29 | 0.03 | 1.00e+00 | 1.00e+00 | 1BCL2L1 |
199 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1MAP2K3 |
200 |
| HALLMARK_ESTROGEN_RESPONSE_EARLY | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1BLVRB |
200 |
| HALLMARK_ESTROGEN_RESPONSE_LATE | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1BLVRB |
200 |
| HALLMARK_MYOGENESIS | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1MYL4 |
200 |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1BPGM |
200 |
| HALLMARK_MTORC1_SIGNALING | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1MAP2K3 |
200 |
| HALLMARK_KRAS_SIGNALING_DN | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1PRODH |
200 |
| HALLMARK_CHOLESTEROL_HOMEOSTASIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
74 |
| HALLMARK_MITOTIC_SPINDLE | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
199 |
| HALLMARK_WNT_BETA_CATENIN_SIGNALING | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
42 |
Dowload full table
KEGG Pathways:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| KEGG_PORPHYRIN_AND_CHLOROPHYLL_METABOLISM | 1.15e-02 | 13.35 | 1.52 | 1.00e+00 | 1.00e+00 | 2ALAS2, BLVRB |
41 |
| KEGG_AMYOTROPHIC_LATERAL_SCLEROSIS_ALS | 1.87e-02 | 10.20 | 1.17 | 1.00e+00 | 1.00e+00 | 2MAP2K3, BCL2L1 |
53 |
| KEGG_ARGININE_AND_PROLINE_METABOLISM | 1.94e-02 | 10.01 | 1.15 | 1.00e+00 | 1.00e+00 | 2ARG1, PRODH |
54 |
| KEGG_PARKINSONS_DISEASE | 9.30e-02 | 4.07 | 0.47 | 1.00e+00 | 1.00e+00 | 2SNCA, UBB |
130 |
| KEGG_PURINE_METABOLISM | 1.29e-01 | 3.32 | 0.39 | 1.00e+00 | 1.00e+00 | 2GMPR, GUK1 |
159 |
| KEGG_THYROID_CANCER | 1.09e-01 | 9.11 | 0.22 | 1.00e+00 | 1.00e+00 | 1NCOA4 |
29 |
| KEGG_GLYCINE_SERINE_AND_THREONINE_METABOLISM | 1.16e-01 | 8.50 | 0.20 | 1.00e+00 | 1.00e+00 | 1ALAS2 |
31 |
| KEGG_PATHWAYS_IN_CANCER | 3.61e-01 | 1.61 | 0.19 | 1.00e+00 | 1.00e+00 | 2NCOA4, BCL2L1 |
325 |
| KEGG_PATHOGENIC_ESCHERICHIA_COLI_INFECTION | 2.00e-01 | 4.64 | 0.11 | 1.00e+00 | 1.00e+00 | 1TUBB1 |
56 |
| KEGG_GLYCOLYSIS_GLUCONEOGENESIS | 2.19e-01 | 4.18 | 0.10 | 1.00e+00 | 1.00e+00 | 1BPGM |
62 |
| KEGG_ADIPOCYTOKINE_SIGNALING_PATHWAY | 2.34e-01 | 3.86 | 0.09 | 1.00e+00 | 1.00e+00 | 1ADIPOR1 |
67 |
| KEGG_PANCREATIC_CANCER | 2.43e-01 | 3.70 | 0.09 | 1.00e+00 | 1.00e+00 | 1BCL2L1 |
70 |
| KEGG_CHRONIC_MYELOID_LEUKEMIA | 2.52e-01 | 3.54 | 0.09 | 1.00e+00 | 1.00e+00 | 1BCL2L1 |
73 |
| KEGG_FC_EPSILON_RI_SIGNALING_PATHWAY | 2.70e-01 | 3.27 | 0.08 | 1.00e+00 | 1.00e+00 | 1MAP2K3 |
79 |
| KEGG_SMALL_CELL_LUNG_CANCER | 2.84e-01 | 3.07 | 0.08 | 1.00e+00 | 1.00e+00 | 1BCL2L1 |
84 |
| KEGG_APOPTOSIS | 2.93e-01 | 2.97 | 0.07 | 1.00e+00 | 1.00e+00 | 1BCL2L1 |
87 |
| KEGG_GAP_JUNCTION | 3.01e-01 | 2.87 | 0.07 | 1.00e+00 | 1.00e+00 | 1TUBB1 |
90 |
| KEGG_GNRH_SIGNALING_PATHWAY | 3.31e-01 | 2.55 | 0.06 | 1.00e+00 | 1.00e+00 | 1MAP2K3 |
101 |
| KEGG_TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY | 3.33e-01 | 2.53 | 0.06 | 1.00e+00 | 1.00e+00 | 1MAP2K3 |
102 |
| KEGG_TIGHT_JUNCTION | 4.08e-01 | 1.95 | 0.05 | 1.00e+00 | 1.00e+00 | 1YBX3 |
132 |
Dowload full table
CHR Positional Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| chr8p21 | 1.40e-02 | 6.38 | 1.25 | 1.00e+00 | 1.00e+00 | 3SLC25A37, BNIP3L, DMTN |
128 |
| chr1p36 | 3.14e-01 | 1.67 | 0.43 | 1.00e+00 | 1.00e+00 | 4RHD, RAP1GAP, SMIM1, PITHD1 |
656 |
| chr19p13 | 5.35e-01 | 1.41 | 0.37 | 1.00e+00 | 1.00e+00 | 4NFIX, FKBP8, TMPRSS9, R3HDM4 |
773 |
| chr17q21 | 2.57e-01 | 1.76 | 0.35 | 1.00e+00 | 1.00e+00 | 3SLC4A1, MYL4, SLC25A39 |
457 |
| chr17p11 | 1.83e-01 | 2.64 | 0.31 | 1.00e+00 | 1.00e+00 | 2UBB, MAP2K3 |
199 |
| chr20q13 | 6.68e-01 | 1.31 | 0.15 | 1.00e+00 | 1.00e+00 | 2FAM210B, TUBB1 |
400 |
| chr7q33 | 1.87e-01 | 5.00 | 0.12 | 1.00e+00 | 1.00e+00 | 1BPGM |
52 |
| chr4q22 | 2.43e-01 | 3.70 | 0.09 | 1.00e+00 | 1.00e+00 | 1SNCA |
70 |
| chr18q12 | 3.17e-01 | 2.68 | 0.07 | 1.00e+00 | 1.00e+00 | 1SLC14A1 |
96 |
| chr6q23 | 3.44e-01 | 2.43 | 0.06 | 1.00e+00 | 1.00e+00 | 1ARG1 |
106 |
| chr20p13 | 3.72e-01 | 2.20 | 0.05 | 1.00e+00 | 1.00e+00 | 1SMOX |
117 |
| chr19q13 | 4.39e-01 | 0.45 | 0.05 | 1.00e+00 | 1.00e+00 | 2KLC3, BLVRB |
1165 |
| chr15q15 | 4.33e-01 | 1.80 | 0.04 | 1.00e+00 | 1.00e+00 | 1CCNDBP1 |
143 |
| chr2q11 | 4.40e-01 | 1.76 | 0.04 | 1.00e+00 | 1.00e+00 | 1FAM178B |
146 |
| chr2q14 | 4.57e-01 | 1.67 | 0.04 | 1.00e+00 | 1.00e+00 | 1GYPC |
154 |
| chr9p13 | 4.90e-01 | 1.51 | 0.04 | 1.00e+00 | 1.00e+00 | 1DCAF12 |
170 |
| chr7q34 | 5.10e-01 | 1.43 | 0.04 | 1.00e+00 | 1.00e+00 | 1MKRN1 |
180 |
| chr10q24 | 5.12e-01 | 1.42 | 0.03 | 1.00e+00 | 1.00e+00 | 1HPS1 |
181 |
| chr10q11 | 5.18e-01 | 1.39 | 0.03 | 1.00e+00 | 1.00e+00 | 1NCOA4 |
184 |
| chr14q24 | 5.31e-01 | 1.34 | 0.03 | 1.00e+00 | 1.00e+00 | 1PLEK2 |
191 |
Dowload full table
Transcription Factor Targets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GATA_C | 5.80e-04 | 6.51 | 2.25 | 5.01e-01 | 6.58e-01 | 6SLC4A1, RHD, BPGM, BLVRB, GFI1B, DMTN |
268 |
| PHB2_TARGET_GENES | 1.03e-03 | 7.19 | 2.20 | 5.01e-01 | 1.00e+00 | 5NFE2, GFI1B, NFIX, FKBP8, GUK1 |
198 |
| GATA3_01 | 2.60e-03 | 5.79 | 1.78 | 5.88e-01 | 1.00e+00 | 5SLC4A1, SNCA, RHD, DMTN, BCL2L1 |
245 |
| GATAAGR_GATA_C | 5.94e-03 | 4.72 | 1.45 | 9.10e-01 | 1.00e+00 | 5SLC4A1, NFE2, RHD, BPGM, DMTN |
299 |
| GATA_Q6 | 7.84e-03 | 5.52 | 1.43 | 9.87e-01 | 1.00e+00 | 4SLC4A1, RHD, BPGM, DMTN |
201 |
| CBFA2T2_TARGET_GENES | 6.43e-03 | 2.61 | 1.27 | 9.10e-01 | 1.00e+00 | 13SELENBP1, NFE2, RHD, ADIPOR1, NCOA4, NFIX, BNIP3L, FKBP8, FAM178B, RNF10, PITHD1, HPS1, GUK1 |
1694 |
| GATA1_03 | 1.54e-02 | 4.49 | 1.17 | 1.00e+00 | 1.00e+00 | 4SLC4A1, NFE2, SNCA, BPGM |
246 |
| GATA1_02 | 1.60e-02 | 4.44 | 1.15 | 1.00e+00 | 1.00e+00 | 4SLC4A1, SNCA, SMOX, BCL2L1 |
249 |
| HMGB1_TARGET_GENES | 1.94e-02 | 2.33 | 1.08 | 1.00e+00 | 1.00e+00 | 11KLC3, GMPR, SMOX, BLVRB, RAP1GAP, NFIX, FAM210B, MAP2K3, PLEK2, RNF10, MKRN1 |
1523 |
| ZNF410_TARGET_GENES | 2.97e-02 | 2.76 | 0.96 | 1.00e+00 | 1.00e+00 | 6MYL4, NFIX, MAP2K3, DMTN, HPS1, GUK1 |
623 |
| GCTNWTTGK_UNKNOWN | 3.20e-02 | 3.55 | 0.92 | 1.00e+00 | 1.00e+00 | 4SNCA, STRADB, UBB, RNF10 |
310 |
| MIF1_01 | 3.64e-02 | 4.36 | 0.86 | 1.00e+00 | 1.00e+00 | 3SLC25A37, UBB, BCL2L1 |
186 |
| CC2D1A_TARGET_GENES | 1.30e-01 | 1.93 | 0.78 | 1.00e+00 | 1.00e+00 | 8GMPR, NFE2, NFIX, FKBP8, FAM178B, MAP2K3, BCL2L1, GUK1 |
1245 |
| CTGCAGY_UNKNOWN | 6.99e-02 | 2.21 | 0.77 | 1.00e+00 | 1.00e+00 | 6GMPR, RAP1GAP, NFIX, DMTN, CCNDBP1, PITHD1 |
779 |
| RYTGCNNRGNAAC_MIF1_01 | 4.93e-02 | 5.92 | 0.69 | 1.00e+00 | 1.00e+00 | 2UBB, BCL2L1 |
90 |
| ZFP91_TARGET_GENES | 2.37e-01 | 1.66 | 0.67 | 1.00e+00 | 1.00e+00 | 8SLC25A37, KLC3, GYPC, MPP1, GFI1B, YBX3, NFIX, FKBP8 |
1442 |
| MZF1_01 | 6.72e-02 | 3.37 | 0.67 | 1.00e+00 | 1.00e+00 | 3BLVRB, YBX3, NFIX |
240 |
| ZNF197_TARGET_GENES | 1.01e-01 | 2.14 | 0.66 | 1.00e+00 | 1.00e+00 | 5NFE2, MYL4, SLC14A1, NFIX, FAM178B |
655 |
| GATA1_04 | 7.18e-02 | 3.27 | 0.65 | 1.00e+00 | 1.00e+00 | 3SLC4A1, NFE2, SLC14A1 |
247 |
| SREBP1_Q6 | 7.18e-02 | 3.27 | 0.65 | 1.00e+00 | 1.00e+00 | 3SLC25A37, SLC4A1, NFIX |
247 |
Dowload full table
GO Biological Processes:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GOBP_ERYTHROCYTE_DEVELOPMENT | 1.50e-07 | 51.27 | 14.77 | 1.12e-03 | 1.12e-03 | 5ALAS2, SLC4A1, BPGM, FAM210B, DMTN |
32 |
| GOBP_MYELOID_CELL_DEVELOPMENT | 5.94e-06 | 22.73 | 6.81 | 2.22e-02 | 4.44e-02 | 5ALAS2, SLC4A1, BPGM, FAM210B, DMTN |
66 |
| GOBP_GDP_METABOLIC_PROCESS | 8.35e-04 | 57.69 | 5.91 | 8.92e-01 | 1.00e+00 | 2MPP1, GUK1 |
11 |
| GOBP_HEME_METABOLIC_PROCESS | 2.96e-04 | 26.56 | 5.01 | 4.43e-01 | 1.00e+00 | 3ALAS2, SLC25A39, BLVRB |
33 |
| GOBP_GMP_METABOLIC_PROCESS | 1.58e-03 | 39.95 | 4.27 | 1.00e+00 | 1.00e+00 | 2MPP1, GUK1 |
15 |
| GOBP_PORPHYRIN_CONTAINING_COMPOUND_METABOLIC_PROCESS | 5.25e-04 | 21.53 | 4.11 | 6.55e-01 | 1.00e+00 | 3ALAS2, SLC25A39, BLVRB |
40 |
| GOBP_REGULATION_OF_KILLING_OF_CELLS_OF_OTHER_ORGANISM | 1.80e-03 | 37.11 | 3.99 | 1.00e+00 | 1.00e+00 | 2ARG1, BCL2L1 |
16 |
| GOBP_ERYTHROCYTE_HOMEOSTASIS | 1.10e-04 | 11.97 | 3.64 | 2.74e-01 | 8.22e-01 | 5ALAS2, SLC4A1, BPGM, FAM210B, DMTN |
121 |
| GOBP_MYELOID_CELL_HOMEOSTASIS | 2.54e-04 | 9.92 | 3.03 | 4.43e-01 | 1.00e+00 | 5ALAS2, SLC4A1, BPGM, FAM210B, DMTN |
145 |
| GOBP_TETRAPYRROLE_METABOLIC_PROCESS | 1.88e-03 | 13.52 | 2.62 | 1.00e+00 | 1.00e+00 | 3ALAS2, SLC25A39, BLVRB |
62 |
| GOBP_GLUTAMINE_FAMILY_AMINO_ACID_CATABOLIC_PROCESS | 5.10e-03 | 20.80 | 2.32 | 1.00e+00 | 1.00e+00 | 2ARG1, PRODH |
27 |
| GOBP_REGULATION_OF_MITOCHONDRIAL_MEMBRANE_POTENTIAL | 2.77e-03 | 11.73 | 2.28 | 1.00e+00 | 1.00e+00 | 3UBB, BNIP3L, BCL2L1 |
71 |
| GOBP_PIGMENT_METABOLIC_PROCESS | 2.88e-03 | 11.56 | 2.25 | 1.00e+00 | 1.00e+00 | 3ALAS2, SLC25A39, BLVRB |
72 |
| GOBP_TETRAPYRROLE_BIOSYNTHETIC_PROCESS | 5.48e-03 | 20.00 | 2.24 | 1.00e+00 | 1.00e+00 | 2ALAS2, SLC25A39 |
28 |
| GOBP_GLYCOLYTIC_PROCESS_THROUGH_FRUCTOSE_6_PHOSPHATE | 5.87e-03 | 19.27 | 2.16 | 1.00e+00 | 1.00e+00 | 2SLC4A1, BPGM |
29 |
| GOBP_NEGATIVE_REGULATION_OF_ATP_METABOLIC_PROCESS | 5.87e-03 | 19.27 | 2.16 | 1.00e+00 | 1.00e+00 | 2SLC4A1, SNCA |
29 |
| GOBP_NEGATIVE_REGULATION_OF_PROTEIN_LOCALIZATION_TO_MEMBRANE | 6.27e-03 | 18.58 | 2.09 | 1.00e+00 | 1.00e+00 | 2DMTN, BCL2L1 |
30 |
| GOBP_REGULATION_OF_AUTOPHAGY_OF_MITOCHONDRION | 6.69e-03 | 17.94 | 2.02 | 1.00e+00 | 1.00e+00 | 2BNIP3L, FBXO7 |
31 |
| GOBP_NEGATIVE_REGULATION_OF_SIGNAL_TRANSDUCTION_IN_ABSENCE_OF_LIGAND | 7.12e-03 | 17.34 | 1.95 | 1.00e+00 | 1.00e+00 | 2STRADB, BCL2L1 |
32 |
| GOBP_IRON_ION_HOMEOSTASIS | 4.74e-03 | 9.61 | 1.88 | 1.00e+00 | 1.00e+00 | 3ALAS2, SLC25A37, NCOA4 |
86 |
Dowload full table
Immunological Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GSE34205_RSV_VS_FLU_INF_INFANT_PBMC_UP | 1.89e-27 | 57.06 | 30.44 | 9.20e-24 | 9.20e-24 | 22ALAS2, SELENBP1, SLC25A37, SLC4A1, MYL4, SNCA, STRADB, SLC14A1, BPGM, SLC25A39, ADIPOR1, RAP1GAP, BNIP3L, DCAF12, FAM210B, FBXO7, PLEK2, CCNDBP1, RNF10, MKRN1, PITHD1, BCL2L1 |
194 |
| GSE34205_HEALTHY_VS_RSV_INF_INFANT_PBMC_DN | 1.78e-20 | 38.62 | 20.01 | 4.33e-17 | 8.66e-17 | 18ALAS2, SELENBP1, SLC4A1, GMPR, MYL4, SNCA, STRADB, ARG1, BPGM, BLVRB, ADIPOR1, MPP1, RAP1GAP, BNIP3L, DCAF12, FBXO7, RNF10, MKRN1 |
200 |
| GSE6269_FLU_VS_STREP_PNEUMO_INF_PBMC_DN | 5.43e-05 | 10.27 | 3.53 | 7.75e-02 | 2.65e-01 | 6SNCA, SLC14A1, ADIPOR1, YBX3, NCOA4, BNIP3L |
172 |
| GSE6269_HEALTHY_VS_STAPH_AUREUS_INF_PBMC_DN | 6.36e-05 | 9.97 | 3.42 | 7.75e-02 | 3.10e-01 | 6SNCA, MPP1, YBX3, BNIP3L, MAP2K3, RNF10 |
177 |
| GSE27786_NKCELL_VS_NEUTROPHIL_DN | 1.24e-04 | 8.78 | 3.02 | 1.21e-01 | 6.03e-01 | 6STRADB, ADIPOR1, DCAF12, DMTN, CCNDBP1, PITHD1 |
200 |
| GSE6269_HEALTHY_VS_STAPH_PNEUMO_INF_PBMC_DN | 5.24e-04 | 8.42 | 2.57 | 4.25e-01 | 1.00e+00 | 5SLC25A37, SNCA, BLVRB, ADIPOR1, BNIP3L |
170 |
| GSE25088_ROSIGLITAZONE_VS_IL4_AND_ROSIGLITAZONE_STIM_STAT6_KO_MACROPHAGE_DAY10_UP | 1.01e-03 | 7.23 | 2.22 | 4.38e-01 | 1.00e+00 | 5SLC25A37, NCOA4, BNIP3L, FKBP8, MKRN1 |
197 |
| GSE27786_LIN_NEG_VS_NKCELL_UP | 1.08e-03 | 7.12 | 2.18 | 4.38e-01 | 1.00e+00 | 5RHD, BLVRB, TUBB1, PITHD1, PRODH |
200 |
| GSE27786_CD4_TCELL_VS_NKCELL_DN | 1.08e-03 | 7.12 | 2.18 | 4.38e-01 | 1.00e+00 | 5SLC25A39, BLVRB, ADIPOR1, MPP1, NFIX |
200 |
| GSE39820_TGFBETA3_IL6_VS_TGFBETA3_IL6_IL23A_TREATED_CD4_TCELL_DN | 1.08e-03 | 7.12 | 2.18 | 4.38e-01 | 1.00e+00 | 5SELENBP1, MYL4, SMOX, YBX3, FBXO7 |
200 |
| GSE369_SOCS3_KO_VS_WT_LIVER_POST_IL6_INJECTION_DN | 1.08e-03 | 7.12 | 2.18 | 4.38e-01 | 1.00e+00 | 5MYL4, STRADB, RHD, SLC14A1, BLVRB |
200 |
| GSE21774_CD56_BRIGHT_VS_DIM_CD62L_POSITIVE_NK_CELL_UP | 1.08e-03 | 7.12 | 2.18 | 4.38e-01 | 1.00e+00 | 5RHD, BPGM, GYPC, BLVRB, GFI1B |
200 |
| GSE6269_FLU_VS_STAPH_AUREUS_INF_PBMC_DN | 4.13e-03 | 6.67 | 1.72 | 9.63e-01 | 1.00e+00 | 4BLVRB, ADIPOR1, MPP1, BNIP3L |
167 |
| GSE2706_2H_VS_8H_R848_STIM_DC_UP | 6.95e-03 | 5.72 | 1.48 | 9.63e-01 | 1.00e+00 | 4BLVRB, MPP1, FAM210B, MAP2K3 |
194 |
| GSE9509_LPS_VS_LPS_AND_IL10_STIM_IL10_KO_MACROPHAGE_20MIN_UP | 6.95e-03 | 5.72 | 1.48 | 9.63e-01 | 1.00e+00 | 4GMPR, GFI1B, FAM210B, SMIM1 |
194 |
| GSE34156_NOD2_LIGAND_VS_TLR1_TLR2_LIGAND_6H_TREATED_MONOCYTE_DN | 7.32e-03 | 5.63 | 1.46 | 9.63e-01 | 1.00e+00 | 4SLC25A37, BLVRB, MPP1, NCOA4 |
197 |
| GSE19198_6H_VS_24H_IL21_TREATED_TCELL_DN | 7.45e-03 | 5.60 | 1.45 | 9.63e-01 | 1.00e+00 | 4SNCA, BLVRB, MPP1, PITHD1 |
198 |
| GSE3039_B2_VS_B1_BCELL_UP | 7.58e-03 | 5.57 | 1.44 | 9.63e-01 | 1.00e+00 | 4ALAS2, SLC25A37, SLC4A1, SNCA |
199 |
| GSE22229_UNTREATED_VS_IMMUNOSUPP_THERAPY_RENAL_TRANSPLANT_PATIENT_PBMC_DN | 7.58e-03 | 5.57 | 1.44 | 9.63e-01 | 1.00e+00 | 4SELENBP1, SLC14A1, SMOX, MKRN1 |
199 |
| GSE21546_WT_VS_ELK1_KO_DP_THYMOCYTES_UP | 7.58e-03 | 5.57 | 1.44 | 9.63e-01 | 1.00e+00 | 4NFE2, SMOX, YBX3, R3HDM4 |
199 |
Top Ranked Transcription Factors for this Gene Expression Program:
| Gene Symbol | TF Rank | DNA Binding Domain | Motif Status | IUPAC PWM | GTEx | DepMap | Decartes |
|---|---|---|---|---|---|---|---|
| NFE2 | 7 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| UBB | 20 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| GFI1B | 24 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| YBX3 | 25 | Yes | Known motif | Monomer or homomultimer | 100 perc ID - in vitro | None | Identical DBD to YBX1. Might also bind RNA. |
| NCOA4 | 26 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| NFIX | 27 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| PLEK2 | 38 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| MKRN1 | 43 | No | ssDNA/RNA binding | Not a DNA binding protein | No motif | None | None |
| CAT | 52 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Protein that breaks down hydrogen peroxide |
| TAL1 | 55 | Yes | Known motif | Obligate heteromer | In vivo/Misc source | Only known motifs are from Transfac or HocoMoco - origin is uncertain | No motif yielded from PBMs or HT-SELEX. Binds DNA as heterodimer with TCF3 (PDB:2YPB and PDB:2YPA). |
| CTCFL | 66 | Yes | Known motif | Monomer or homomultimer | In vivo/Misc source | None | None |
| LYL1 | 69 | Yes | Inferred motif | Obligate heteromer | In vivo/Misc source | None | TAL1, TAL2 and LYL are very similar and have a basic region with a bulky tryptophan inserting into the cluster of the basic residues. Binds DNA as a heterodimer with TCF3 (PDB:2YPB and PDB:2YPA). |
| FOXO4 | 90 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| HSF5 | 94 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| FANK1 | 101 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Binds JUN to control its activity (PMID: 20978819), but there is no evidence that it binds DNA itself |
| PKHD1L1 | 108 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Membrane receptor (PMID: 12620974) |
| KLF8 | 115 | Yes | Known motif | Monomer or homomultimer | 100 perc ID - in vitro | None | None |
| CREG1 | 127 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Regulates transcription in tethering assays (PMID: 9710587). Unlikely to be a TF, based on the lack of a canonical DBD |
| ELF4 | 131 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| PIM1 | 149 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Affects transcription by phosphorylating TFs (PMID: 18593906) |
QQ Plot showing correlations with other GEPs in this dataset, calculated by Spearman correlation:
Interactive QQ-plot of gene loadings:
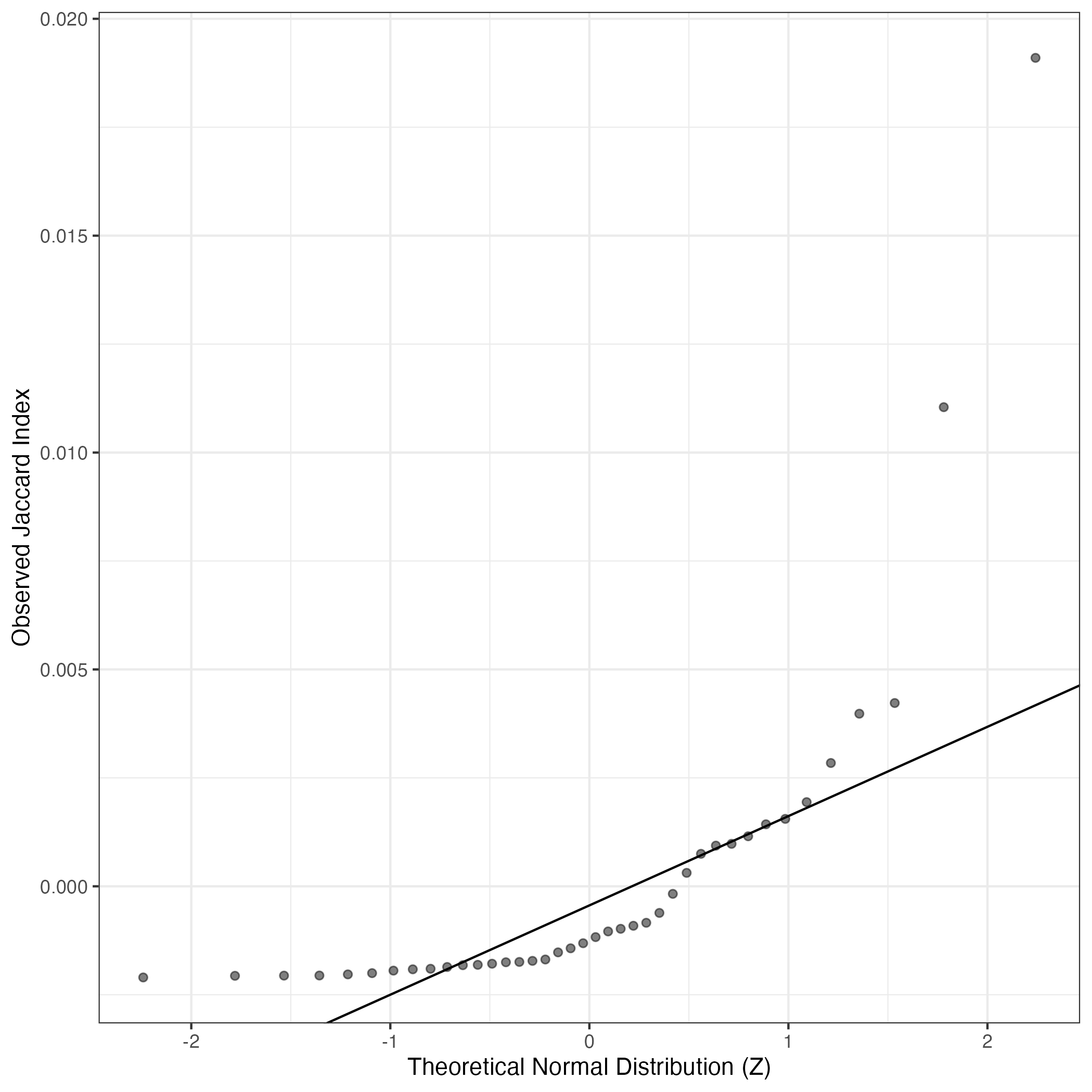
A similar QQ-plot as above, but only for instances where the H value is e.g. > 25, i.e. we are confident that the expression program is active above noise. Agreemenet between these binary vectors is tested using the Jaccard Index, with the P-values calculated by an exact test:
Interactive QQ-plot:
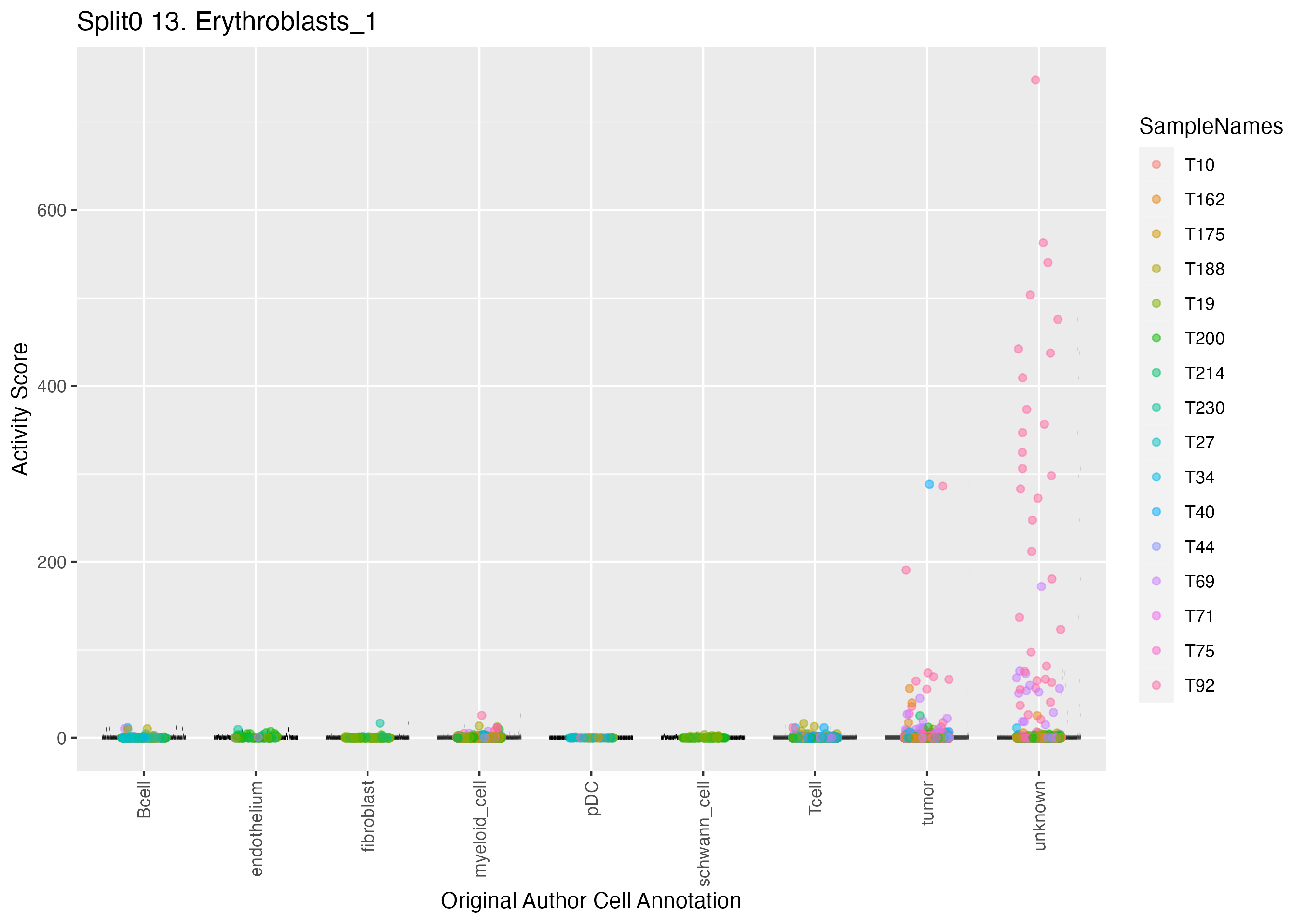
Singler cell type annotations for the top 50 cells on this program.
| Cell ID | Singler label | Singler Delta | Activity Score | Top Singler Raw Scores |
|---|---|---|---|---|
| T92_GATGAAACAAGCGATG.1 | Erythroblast | 0.08 | 747.79 | Raw ScoresBM: 0.14, Erythroblast: 0.14, Pro-Myelocyte: 0.11, MEP: 0.1, CMP: 0.08, Pro-B_cell_CD34+: 0.08, Monocyte: 0.08, Pre-B_cell_CD34-: 0.07, B_cell:immature: 0.07, Endothelial_cells:HUVEC:Borrelia_burgdorferi: 0.07 |
| T92_GACGTGCGTAAACGCG.1 | Erythroblast | 0.05 | 562.69 | Raw ScoresErythroblast: 0.14, BM: 0.13, Neurons:adrenal_medulla_cell_line: 0.13, MEP: 0.12, Neuroepithelial_cell:ESC-derived: 0.12, Pro-Myelocyte: 0.12, CMP: 0.11, Tissue_stem_cells:CD326-CD56+: 0.11, Pro-B_cell_CD34+: 0.11, Astrocyte:Embryonic_stem_cell-derived: 0.11 |
| T92_GCAATCACAGGCAGTA.1 | BM | 0.07 | 540.10 | Raw ScoresBM: 0.22, B_cell:immature: 0.19, Pro-B_cell_CD34+: 0.19, Pro-Myelocyte: 0.19, Erythroblast: 0.19, Pre-B_cell_CD34-: 0.18, MEP: 0.18, T_cell:CD8+: 0.18, CMP: 0.18, B_cell:Naive: 0.18 |
| T92_GTTTCTAAGCCCGAAA.1 | Erythroblast | 0.07 | 503.40 | Raw ScoresErythroblast: 0.15, BM: 0.14, MEP: 0.12, Pro-Myelocyte: 0.12, CMP: 0.1, T_cell:CD8+_naive: 0.1, T_cell:CD8+_effector_memory: 0.1, T_cell:CD8+: 0.09, Pre-B_cell_CD34-: 0.09, T_cell:CD8+_Central_memory: 0.09 |
| T92_AGCTCTCTCTCTGCTG.1 | Erythroblast | 0.06 | 475.58 | Raw ScoresErythroblast: 0.17, BM: 0.17, MEP: 0.16, Pro-Myelocyte: 0.15, Pro-B_cell_CD34+: 0.14, CMP: 0.14, B_cell:immature: 0.13, Pre-B_cell_CD34-: 0.13, GMP: 0.13, HSC_CD34+: 0.13 |
| T92_AGTGTCACAAGTAATG.1 | Erythroblast | 0.06 | 442.05 | Raw ScoresErythroblast: 0.17, BM: 0.16, MEP: 0.15, Pro-Myelocyte: 0.14, Pro-B_cell_CD34+: 0.13, T_cell:CD8+_naive: 0.13, CMP: 0.13, T_cell:CD8+_effector_memory: 0.13, T_cell:CD8+_Central_memory: 0.13, T_cell:CD4+_central_memory: 0.13 |
| T92_GCCAAATAGGCGTACA.1 | BM | 0.07 | 437.32 | Raw ScoresBM: 0.15, Erythroblast: 0.15, MEP: 0.13, Pro-Myelocyte: 0.13, CMP: 0.12, Pro-B_cell_CD34+: 0.12, HSC_CD34+: 0.12, GMP: 0.11, Pre-B_cell_CD34-: 0.1, Endothelial_cells:HUVEC:Borrelia_burgdorferi: 0.1 |
| T92_CGTCCATGTGAAAGAG.1 | BM | 0.07 | 409.14 | Raw ScoresErythroblast: 0.16, BM: 0.16, MEP: 0.15, Pro-Myelocyte: 0.14, CMP: 0.13, Pro-B_cell_CD34+: 0.13, Endothelial_cells:HUVEC:FPV-infected: 0.12, HSC_CD34+: 0.12, GMP: 0.12, Endothelial_cells:HUVEC:Borrelia_burgdorferi: 0.12 |
| T92_GTGAAGGGTAAGGATT.1 | Erythroblast | 0.06 | 373.33 | Raw ScoresErythroblast: 0.17, BM: 0.17, Pro-Myelocyte: 0.16, MEP: 0.15, CMP: 0.14, Pro-B_cell_CD34+: 0.14, Neurons:adrenal_medulla_cell_line: 0.13, Pre-B_cell_CD34-: 0.13, T_cell:CD8+_naive: 0.13, Myelocyte: 0.13 |
| T92_ACGAGCCAGTGACATA.1 | Erythroblast | 0.05 | 356.50 | Raw ScoresErythroblast: 0.19, Neurons:adrenal_medulla_cell_line: 0.18, BM: 0.18, MEP: 0.18, Pro-Myelocyte: 0.17, CMP: 0.17, Pro-B_cell_CD34+: 0.17, Neuroepithelial_cell:ESC-derived: 0.17, iPS_cells:fibroblast-derived:Direct_del._reprog: 0.17, Neurons:Schwann_cell: 0.17 |
| T92_AGATCTGGTTCACCTC.1 | Erythroblast | 0.05 | 346.91 | Raw ScoresErythroblast: 0.19, Neurons:adrenal_medulla_cell_line: 0.17, BM: 0.17, MEP: 0.17, Astrocyte:Embryonic_stem_cell-derived: 0.16, Neuroepithelial_cell:ESC-derived: 0.16, Pro-Myelocyte: 0.16, Neurons:Schwann_cell: 0.16, iPS_cells:adipose_stem_cell-derived:lentiviral: 0.16, iPS_cells:fibroblast-derived:Retroviral_transf: 0.16 |
| T92_ACGAGGACACTGTTAG.1 | Neurons:adrenal_medulla_cell_line | 0.06 | 324.47 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.19, MEP: 0.17, CMP: 0.17, Erythroblast: 0.17, BM: 0.17, Pro-B_cell_CD34+: 0.16, Neuroepithelial_cell:ESC-derived: 0.16, Pro-Myelocyte: 0.16, Endothelial_cells:HUVEC:FPV-infected: 0.16, Endothelial_cells:HUVEC:Borrelia_burgdorferi: 0.16 |
| T92_TGCGTGGCATCTACGA.1 | Erythroblast | 0.06 | 306.02 | Raw ScoresBM: 0.17, Erythroblast: 0.17, Pro-Myelocyte: 0.15, MEP: 0.15, T_cell:CD8+_naive: 0.13, Pro-B_cell_CD34+: 0.13, CMP: 0.13, T_cell:CD8+_Central_memory: 0.13, T_cell:CD8+_effector_memory: 0.13, HSC_CD34+: 0.13 |
| T92_CGTCACTTCGAACGGA.1 | Erythroblast | 0.06 | 297.93 | Raw ScoresErythroblast: 0.2, Neurons:adrenal_medulla_cell_line: 0.19, BM: 0.19, Pro-Myelocyte: 0.19, MEP: 0.19, Pro-B_cell_CD34+: 0.18, Neuroepithelial_cell:ESC-derived: 0.18, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.18, iPS_cells:adipose_stem_cell-derived:lentiviral: 0.18, iPS_cells:fibroblast-derived:Retroviral_transf: 0.17 |
| T40_CAGGTGCTCGACAGCC.1 | Erythroblast | 0.12 | 288.34 | Raw ScoresMEP: 0.4, Erythroblast: 0.39, CMP: 0.36, Pro-Myelocyte: 0.35, GMP: 0.35, Pro-B_cell_CD34+: 0.35, BM: 0.34, HSC_CD34+: 0.33, T_cell:gamma-delta: 0.32, NK_cell:IL2: 0.31 |
| T92_TGGTTCCCAATCACAC.1 | Neurons:adrenal_medulla_cell_line | 0.11 | 286.14 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.25, Neuroepithelial_cell:ESC-derived: 0.23, iPS_cells:fibroblast-derived:Retroviral_transf: 0.23, iPS_cells:fibroblast-derived:Direct_del._reprog: 0.22, Erythroblast: 0.22, iPS_cells:PDB_1lox-21Puro-20: 0.22, iPS_cells:PDB_1lox-21Puro-26: 0.22, iPS_cells:PDB_1lox-17Puro-10: 0.22, iPS_cells:PDB_1lox-17Puro-5: 0.22, Embryonic_stem_cells: 0.22 |
| T92_TAAGCGTAGTTCCACA.1 | BM | 0.05 | 282.92 | Raw ScoresBM: 0.17, Erythroblast: 0.15, Pro-Myelocyte: 0.15, Pre-B_cell_CD34-: 0.15, Pro-B_cell_CD34+: 0.15, Monocyte:CD16-: 0.15, NK_cell:CD56hiCD62L+: 0.14, Monocyte: 0.14, B_cell:immature: 0.14, T_cell:CD4+: 0.14 |
| T92_GTACTCCCAATAGCGG.1 | BM | 0.04 | 272.46 | Raw ScoresErythroblast: 0.18, BM: 0.17, MEP: 0.17, Pro-Myelocyte: 0.16, CMP: 0.16, Pro-B_cell_CD34+: 0.16, HSC_CD34+: 0.15, Endothelial_cells:HUVEC:FPV-infected: 0.14, GMP: 0.14, Monocyte:CD16+: 0.14 |
| T92_GCTGGGTCAGCGTCCA.1 | Erythroblast | 0.05 | 247.40 | Raw ScoresErythroblast: 0.15, BM: 0.15, Pro-Myelocyte: 0.14, MEP: 0.12, Pro-B_cell_CD34+: 0.12, CMP: 0.12, Neutrophil:GM-CSF_IFNg: 0.11, Monocyte: 0.11, Neurons:adrenal_medulla_cell_line: 0.11, Pre-B_cell_CD34-: 0.11 |
| T92_CTGAAACAGTGGGTTG.1 | BM | 0.04 | 211.85 | Raw ScoresBM: 0.17, Pro-Myelocyte: 0.17, Endothelial_cells:HUVEC:Borrelia_burgdorferi: 0.16, Erythroblast: 0.16, Pro-B_cell_CD34+: 0.16, Endothelial_cells:HUVEC:IFNg: 0.16, CMP: 0.16, MEP: 0.16, Endothelial_cells:HUVEC:FPV-infected: 0.15, Neurons:Schwann_cell: 0.15 |
| T92_TGTATTCCAGCTGTAT.1 | Neurons:adrenal_medulla_cell_line | 0.12 | 190.64 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Neuroepithelial_cell:ESC-derived: 0.26, Astrocyte:Embryonic_stem_cell-derived: 0.24, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.23, iPS_cells:adipose_stem_cell-derived:lentiviral: 0.23, Tissue_stem_cells:CD326-CD56+: 0.23, Embryonic_stem_cells: 0.23, iPS_cells:PDB_1lox-17Puro-10: 0.23, iPS_cells:PDB_2lox-22: 0.23, iPS_cells:PDB_1lox-21Puro-20: 0.23 |
| T92_TACTTGTGTCAGAAGC.1 | BM | 0.04 | 180.67 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.19, MEP: 0.18, BM: 0.18, Erythroblast: 0.18, Pro-Myelocyte: 0.17, Pro-B_cell_CD34+: 0.17, Neuroepithelial_cell:ESC-derived: 0.17, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.17, iPS_cells:fibroblast-derived:Retroviral_transf: 0.17, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.17 |
| T69_ACGATGTTCTTCTGGC.1 | Neurons:adrenal_medulla_cell_line | 0.12 | 171.97 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Neuroepithelial_cell:ESC-derived: 0.27, Astrocyte:Embryonic_stem_cell-derived: 0.25, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.25, iPS_cells:PDB_2lox-22: 0.25, Embryonic_stem_cells: 0.25, Neurons:ES_cell-derived_neural_precursor: 0.25, iPS_cells:PDB_1lox-21Puro-20: 0.24, iPS_cells:PDB_1lox-17Puro-5: 0.24, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.24 |
| T92_CTAGTGACAGTCAGAG.1 | Neurons:adrenal_medulla_cell_line | 0.04 | 136.93 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.18, MEP: 0.16, Erythroblast: 0.16, Astrocyte:Embryonic_stem_cell-derived: 0.16, BM: 0.16, iPS_cells:fibroblast-derived:Retroviral_transf: 0.16, Neuroepithelial_cell:ESC-derived: 0.16, Pro-B_cell_CD34+: 0.15, Tissue_stem_cells:CD326-CD56+: 0.15, CMP: 0.15 |
| T92_GTCACGGGTTGGTAAA.1 | Erythroblast | 0.06 | 123.05 | Raw ScoresErythroblast: 0.2, BM: 0.19, MEP: 0.19, Neurons:adrenal_medulla_cell_line: 0.18, Pro-Myelocyte: 0.18, CMP: 0.18, Pro-B_cell_CD34+: 0.17, Neuroepithelial_cell:ESC-derived: 0.17, Astrocyte:Embryonic_stem_cell-derived: 0.17, Smooth_muscle_cells:umbilical_vein: 0.17 |
| T92_ACGGAGATCTGAGGGA.1 | T_cell:CD8+_naive | 0.05 | 97.36 | Raw ScoresT_cell:CD4+_central_memory: 0.21, T_cell:CD4+_effector_memory: 0.21, T_cell:CD8+_naive: 0.21, T_cell:CD4+_Naive: 0.21, T_cell:CD8+_effector_memory: 0.21, T_cell:CD8+_Central_memory: 0.21, NK_cell: 0.2, T_cell:CD4+: 0.2, T_cell:gamma-delta: 0.2, T_cell:CD8+_effector_memory_RA: 0.2 |
| T92_TTTGGTTGTATTACCG.1 | T_cell:CD4+_central_memory | 0.10 | 81.58 | Raw ScoresT_cell:CD4+_central_memory: 0.28, T_cell:CD4+_Naive: 0.27, T_cell:CD4+_effector_memory: 0.27, T_cell:CD4+: 0.27, T_cell:CD8+: 0.26, T_cell:CD8+_effector_memory: 0.25, NK_cell: 0.25, Pre-B_cell_CD34-: 0.25, T_cell:CD8+_naive: 0.25, T_cell:gamma-delta: 0.25 |
| T69_CAGATCAGTCATCCCT.1 | Neurons:adrenal_medulla_cell_line | 0.14 | 75.86 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Neuroepithelial_cell:ESC-derived: 0.29, iPS_cells:PDB_1lox-17Puro-5: 0.27, iPS_cells:PDB_1lox-17Puro-10: 0.27, iPS_cells:PDB_1lox-21Puro-20: 0.27, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.27, Astrocyte:Embryonic_stem_cell-derived: 0.27, iPS_cells:PDB_1lox-21Puro-26: 0.27, iPS_cells:PDB_2lox-22: 0.26, Neurons:ES_cell-derived_neural_precursor: 0.26 |
| T92_CAGCATAAGACATAAC.1 | Neurons:adrenal_medulla_cell_line | 0.12 | 75.40 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.27, Neuroepithelial_cell:ESC-derived: 0.22, Astrocyte:Embryonic_stem_cell-derived: 0.22, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.2, iPS_cells:PDB_2lox-22: 0.2, iPS_cells:fibroblast-derived:Retroviral_transf: 0.2, Embryonic_stem_cells: 0.2, iPS_cells:PDB_2lox-21: 0.2, iPS_cells:PDB_1lox-17Puro-5: 0.2, iPS_cells:PDB_2lox-5: 0.2 |
| T92_TACTTACAGGTAGCCA.1 | Neurons:adrenal_medulla_cell_line | 0.14 | 73.73 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Neuroepithelial_cell:ESC-derived: 0.28, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.26, Astrocyte:Embryonic_stem_cell-derived: 0.26, iPS_cells:PDB_2lox-22: 0.26, iPS_cells:adipose_stem_cell-derived:lentiviral: 0.26, iPS_cells:iPS:minicircle-derived: 0.26, iPS_cells:PDB_2lox-17: 0.26, iPS_cells:PDB_1lox-17Puro-10: 0.26, iPS_cells:PDB_1lox-21Puro-20: 0.26 |
| T69_CTCGTACAGGTGCTTT.1 | Neurons:adrenal_medulla_cell_line | 0.08 | 73.11 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.22, Neuroepithelial_cell:ESC-derived: 0.19, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.18, Astrocyte:Embryonic_stem_cell-derived: 0.18, iPS_cells:PDB_1lox-17Puro-5: 0.18, iPS_cells:PDB_2lox-17: 0.18, iPS_cells:PDB_1lox-17Puro-10: 0.18, iPS_cells:fibroblast-derived:Retroviral_transf: 0.18, iPS_cells:PDB_2lox-22: 0.18, iPS_cells:PDB_1lox-21Puro-20: 0.18 |
| T92_GCATGTAAGACTAGGC.1 | Neurons:adrenal_medulla_cell_line | 0.19 | 69.21 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.37, Neuroepithelial_cell:ESC-derived: 0.3, Astrocyte:Embryonic_stem_cell-derived: 0.29, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.26, iPS_cells:PDB_1lox-17Puro-10: 0.26, iPS_cells:PDB_1lox-21Puro-20: 0.26, iPS_cells:PDB_2lox-22: 0.26, iPS_cells:PDB_2lox-21: 0.26, Embryonic_stem_cells: 0.26, iPS_cells:PDB_1lox-17Puro-5: 0.26 |
| T69_CACCTTGAGCTCAACT.1 | Neurons:adrenal_medulla_cell_line | 0.14 | 68.14 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Neuroepithelial_cell:ESC-derived: 0.3, Astrocyte:Embryonic_stem_cell-derived: 0.29, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.27, iPS_cells:PDB_1lox-17Puro-5: 0.27, iPS_cells:PDB_1lox-21Puro-20: 0.27, iPS_cells:PDB_1lox-21Puro-26: 0.27, iPS_cells:PDB_2lox-22: 0.27, iPS_cells:PDB_1lox-17Puro-10: 0.26, iPS_cells:PDB_2lox-17: 0.26 |
| T92_AACTCAGAGAGCTGCA.1 | Neurons:adrenal_medulla_cell_line | 0.11 | 66.65 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.28, Neuroepithelial_cell:ESC-derived: 0.23, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.23, Astrocyte:Embryonic_stem_cell-derived: 0.23, iPS_cells:PDB_2lox-22: 0.22, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.22, iPS_cells:adipose_stem_cell-derived:lentiviral: 0.22, iPS_cells:PDB_2lox-17: 0.22, iPS_cells:PDB_2lox-5: 0.22, iPS_cells:PDB_1lox-21Puro-20: 0.22 |
| T92_GCTTCCAAGATGCGAC.1 | Neurons:adrenal_medulla_cell_line | 0.13 | 66.37 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.32, Neuroepithelial_cell:ESC-derived: 0.27, Astrocyte:Embryonic_stem_cell-derived: 0.27, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.25, iPS_cells:PDB_2lox-22: 0.25, iPS_cells:adipose_stem_cell-derived:lentiviral: 0.25, iPS_cells:PDB_2lox-17: 0.25, iPS_cells:PDB_1lox-17Puro-10: 0.25, Tissue_stem_cells:CD326-CD56+: 0.25, iPS_cells:iPS:minicircle-derived: 0.25 |
| T92_GCCTCTATCGCGTTTC.1 | Neurons:adrenal_medulla_cell_line | 0.15 | 64.99 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.35, Neuroepithelial_cell:ESC-derived: 0.29, Astrocyte:Embryonic_stem_cell-derived: 0.27, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.26, iPS_cells:adipose_stem_cell-derived:lentiviral: 0.26, Embryonic_stem_cells: 0.26, iPS_cells:PDB_2lox-22: 0.26, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.26, iPS_cells:PDB_2lox-21: 0.26, Tissue_stem_cells:CD326-CD56+: 0.26 |
| T92_ACTGCTCAGTAATCCC.1 | Neurons:adrenal_medulla_cell_line | 0.17 | 64.47 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.35, Neuroepithelial_cell:ESC-derived: 0.29, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.27, iPS_cells:PDB_2lox-22: 0.27, iPS_cells:PDB_2lox-21: 0.27, Astrocyte:Embryonic_stem_cell-derived: 0.27, iPS_cells:PDB_2lox-17: 0.26, Tissue_stem_cells:CD326-CD56+: 0.26, iPS_cells:PDB_2lox-5: 0.26, iPS_cells:PDB_1lox-21Puro-20: 0.26 |
| T92_GACTAACGTTCCCGAG.1 | Neurons:adrenal_medulla_cell_line | 0.11 | 63.06 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.25, Neuroepithelial_cell:ESC-derived: 0.22, Astrocyte:Embryonic_stem_cell-derived: 0.21, iPS_cells:fibroblast-derived:Retroviral_transf: 0.2, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.2, iPS_cells:fibroblast-derived:Direct_del._reprog: 0.19, iPS_cells:PDB_2lox-22: 0.19, iPS_cells:PDB_1lox-17Puro-5: 0.19, iPS_cells:PDB_1lox-21Puro-20: 0.19, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.19 |
| T69_GACGGCTGTGGTCCGT.1 | Neurons:adrenal_medulla_cell_line | 0.12 | 59.63 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Neuroepithelial_cell:ESC-derived: 0.27, Astrocyte:Embryonic_stem_cell-derived: 0.26, Neurons:ES_cell-derived_neural_precursor: 0.24, Embryonic_stem_cells: 0.24, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.24, iPS_cells:PDB_2lox-22: 0.24, iPS_cells:PDB_1lox-21Puro-20: 0.24, iPS_cells:PDB_1lox-17Puro-5: 0.24, iPS_cells:PDB_2lox-5: 0.24 |
| T92_GACAGAGGTAAGTTCC.1 | B_cell:immature | 0.07 | 56.55 | Raw ScoresB_cell:immature: 0.21, B_cell:Naive: 0.21, B_cell:Memory: 0.21, B_cell: 0.2, Monocyte:CD16+: 0.2, Pro-B_cell_CD34+: 0.2, B_cell:Plasma_cell: 0.2, Monocyte:CD16-: 0.2, Monocyte:CD14+: 0.2, Pre-B_cell_CD34-: 0.2 |
| T69_TCTCTAAGTGGTGTAG.1 | Neurons:adrenal_medulla_cell_line | 0.13 | 56.13 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.3, Neuroepithelial_cell:ESC-derived: 0.27, Astrocyte:Embryonic_stem_cell-derived: 0.26, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.25, iPS_cells:PDB_1lox-17Puro-5: 0.25, Embryonic_stem_cells: 0.25, iPS_cells:PDB_1lox-21Puro-20: 0.25, iPS_cells:PDB_1lox-17Puro-10: 0.25, Neurons:ES_cell-derived_neural_precursor: 0.24, iPS_cells:PDB_2lox-22: 0.24 |
| T162_AGTGACTCACCAAAGG-1 | Neurons:adrenal_medulla_cell_line | 0.08 | 56.10 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.23, Neuroepithelial_cell:ESC-derived: 0.2, Astrocyte:Embryonic_stem_cell-derived: 0.2, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.19, iPS_cells:PDB_1lox-21Puro-26: 0.19, iPS_cells:PDB_1lox-21Puro-20: 0.19, iPS_cells:PDB_1lox-17Puro-5: 0.19, iPS_cells:PDB_2lox-22: 0.19, Embryonic_stem_cells: 0.19, iPS_cells:iPS:minicircle-derived: 0.19 |
| T92_CTAGAGTTCAGTGCAT.1 | Neurons:adrenal_medulla_cell_line | 0.09 | 55.23 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.27, Neuroepithelial_cell:ESC-derived: 0.24, iPS_cells:PDB_2lox-17: 0.23, iPS_cells:PDB_2lox-22: 0.23, iPS_cells:skin_fibroblast-derived: 0.23, iPS_cells:iPS:minicircle-derived: 0.23, iPS_cells:PDB_2lox-21: 0.23, iPS_cells:adipose_stem_cell-derived:minicircle-derived: 0.23, iPS_cells:adipose_stem_cell-derived:lentiviral: 0.23, iPS_cells:PDB_1lox-21Puro-20: 0.23 |
| T92_CCGTGGAAGCGTAATA.1 | T_cell:CD8+_effector_memory | 0.08 | 54.92 | Raw ScoresT_cell:CD8+_effector_memory: 0.23, T_cell:CD8+_naive: 0.22, T_cell:CD4+_central_memory: 0.22, T_cell:CD4+_Naive: 0.22, T_cell:CD8+: 0.22, T_cell:CD8+_Central_memory: 0.22, T_cell:CD4+_effector_memory: 0.21, T_cell:CD4+: 0.21, T_cell:CD8+_effector_memory_RA: 0.21, T_cell:gamma-delta: 0.21 |
| T69_ATTATCCCAAGAGGCT.1 | Neurons:adrenal_medulla_cell_line | 0.14 | 53.38 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.33, Neuroepithelial_cell:ESC-derived: 0.3, Astrocyte:Embryonic_stem_cell-derived: 0.29, iPS_cells:PDB_2lox-22: 0.28, iPS_cells:PDB_1lox-17Puro-5: 0.28, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.28, iPS_cells:PDB_1lox-21Puro-20: 0.28, iPS_cells:PDB_2lox-17: 0.28, iPS_cells:PDB_2lox-21: 0.28, iPS_cells:PDB_1lox-21Puro-26: 0.28 |
| T69_CACACAATCTAACCGA.1 | Neurons:adrenal_medulla_cell_line | 0.13 | 51.99 | Raw ScoresNeurons:adrenal_medulla_cell_line: 0.32, Neuroepithelial_cell:ESC-derived: 0.28, Astrocyte:Embryonic_stem_cell-derived: 0.27, iPS_cells:PDB_2lox-5: 0.25, iPS_cells:CRL2097_foreskin-derived:undiff.: 0.25, Neurons:ES_cell-derived_neural_precursor: 0.25, iPS_cells:PDB_2lox-22: 0.25, iPS_cells:PDB_2lox-17: 0.25, iPS_cells:PDB_2lox-21: 0.25, iPS_cells:PDB_1lox-17Puro-5: 0.25 |
| T69_AGAGCTTCACCGATAT.1 | Smooth_muscle_cells:vascular:IL-17 | 0.14 | 50.55 | Raw ScoresFibroblasts:breast: 0.28, Smooth_muscle_cells:vascular:IL-17: 0.28, Chondrocytes:MSC-derived: 0.28, Smooth_muscle_cells:vascular: 0.28, iPS_cells:adipose_stem_cells: 0.27, Tissue_stem_cells:BM_MSC:TGFb3: 0.27, Tissue_stem_cells:BM_MSC: 0.27, Tissue_stem_cells:BM_MSC:BMP2: 0.26, Fibroblasts:foreskin: 0.26, Smooth_muscle_cells:bronchial: 0.26 |
Below shows the significant enrichments of this GEP for literature curated gene lists
This data was procured from existing single cell RNA-seq maps of neuroblastoma or related relevant data.
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by the P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Inflammatory CAF
These marker genes were curated across cancer subtypes in multiple organ systems as reviewed in Lavie et. al. (PMID 35883004) and contain inflammatory specific CAF genes:
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.40e-05
Mean rank of genes in gene set: 3242.96
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| C7 | 0.0000074 | 448 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CFB | -0.0000697 | 1243 | GTEx | DepMap | Descartes | 0.02 | 0.81 |
| CXCL14 | -0.0000707 | 1262 | GTEx | DepMap | Descartes | 0.02 | 1.93 |
| IL33 | -0.0000788 | 1359 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| LIF | -0.0000840 | 1435 | GTEx | DepMap | Descartes | 0.06 | 1.62 |
| SCARA5 | -0.0000938 | 1570 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PDGFD | -0.0001114 | 1841 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CFD | -0.0001120 | 1845 | GTEx | DepMap | Descartes | 0.06 | 5.95 |
| IL10 | -0.0001223 | 2004 | GTEx | DepMap | Descartes | 0.02 | 1.19 |
| CXCL12 | -0.0001308 | 2123 | GTEx | DepMap | Descartes | 0.02 | 0.59 |
| IL1R1 | -0.0001541 | 2458 | GTEx | DepMap | Descartes | 0.04 | 0.81 |
| IGFBP6 | -0.0001696 | 2697 | GTEx | DepMap | Descartes | 0.02 | 0.93 |
| PDPN | -0.0001719 | 2729 | GTEx | DepMap | Descartes | 0.04 | 1.35 |
| IGF1 | -0.0001753 | 2782 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CXCL2 | -0.0002246 | 3435 | GTEx | DepMap | Descartes | 1.96 | 172.68 |
| CCL2 | -0.0002319 | 3540 | GTEx | DepMap | Descartes | 0.04 | 3.37 |
| PDGFRB | -0.0002331 | 3565 | GTEx | DepMap | Descartes | 0.09 | 1.51 |
| C3 | -0.0002368 | 3617 | GTEx | DepMap | Descartes | 0.26 | 4.80 |
| SCARA3 | -0.0002723 | 4147 | GTEx | DepMap | Descartes | 0.02 | 0.41 |
| GPX3 | -0.0003169 | 4825 | GTEx | DepMap | Descartes | 0.11 | 7.66 |
| SOD2 | -0.0003645 | 5511 | GTEx | DepMap | Descartes | 0.74 | 5.19 |
| SERPING1 | -0.0004112 | 6227 | GTEx | DepMap | Descartes | 0.09 | 3.42 |
| PDGFRA | -0.0004159 | 6299 | GTEx | DepMap | Descartes | 0.04 | 0.57 |
| RGMA | -0.0004446 | 6720 | GTEx | DepMap | Descartes | 0.02 | 0.14 |
| HGF | -0.0004922 | 7392 | GTEx | DepMap | Descartes | 0.02 | 0.66 |
Myofibroblastic CAF
These marker genes were curated across cancer subtypes in multiple organ systems as reviewed in Lavie et. al. (PMID 35883004) and contain myofibroblastic specific CAF genes:
Wilcoxon ranksum test P-value for gene set overrepresentation: 3.34e-05
Mean rank of genes in gene set: 4215.27
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| POSTN | 0.0000366 | 384 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TNC | -0.0000163 | 557 | GTEx | DepMap | Descartes | 0.04 | 1.27 |
| COL10A1 | -0.0000285 | 716 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| IGFBP3 | -0.0000319 | 773 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PGF | -0.0000652 | 1173 | GTEx | DepMap | Descartes | 0.23 | 6.86 |
| ITGA7 | -0.0000888 | 1498 | GTEx | DepMap | Descartes | 0.04 | 0.64 |
| COL13A1 | -0.0001192 | 1949 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| THBS2 | -0.0001405 | 2264 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| COL5A1 | -0.0001473 | 2362 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ACTG2 | -0.0001480 | 2378 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| BGN | -0.0001520 | 2426 | GTEx | DepMap | Descartes | 0.02 | 0.89 |
| VEGFA | -0.0001546 | 2466 | GTEx | DepMap | Descartes | 0.21 | 2.06 |
| LUM | -0.0001580 | 2511 | GTEx | DepMap | Descartes | 0.02 | 1.43 |
| COL11A1 | -0.0001646 | 2620 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| COL1A1 | -0.0001661 | 2633 | GTEx | DepMap | Descartes | 0.15 | 2.48 |
| DCN | -0.0001710 | 2719 | GTEx | DepMap | Descartes | 0.11 | 1.53 |
| FN1 | -0.0001780 | 2821 | GTEx | DepMap | Descartes | 0.02 | 0.23 |
| COL8A1 | -0.0001926 | 3012 | GTEx | DepMap | Descartes | 0.09 | 1.47 |
| COL3A1 | -0.0002008 | 3125 | GTEx | DepMap | Descartes | 0.17 | 3.05 |
| COL1A2 | -0.0002031 | 3151 | GTEx | DepMap | Descartes | 0.17 | 2.79 |
| TGFB2 | -0.0002167 | 3329 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| WNT5A | -0.0002203 | 3384 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| COL15A1 | -0.0002266 | 3465 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MMP11 | -0.0002279 | 3486 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| COL12A1 | -0.0002472 | 3784 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| COL5A2 | -0.0002547 | 3892 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| COL4A1 | -0.0002596 | 3967 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ACTA2 | -0.0002708 | 4133 | GTEx | DepMap | Descartes | 0.06 | 7.06 |
| MYH11 | -0.0002781 | 4230 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MYLK | -0.0002805 | 4268 | GTEx | DepMap | Descartes | 0.06 | 0.63 |
| THBS1 | -0.0002878 | 4398 | GTEx | DepMap | Descartes | 0.06 | 0.81 |
| TAGLN | -0.0002907 | 4439 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MYL9 | -0.0002945 | 4501 | GTEx | DepMap | Descartes | 0.04 | 1.50 |
| COL14A1 | -0.0003009 | 4605 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MMP2 | -0.0003083 | 4715 | GTEx | DepMap | Descartes | 0.09 | 1.79 |
| HOPX | -0.0003179 | 4844 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MEF2C | -0.0003308 | 5028 | GTEx | DepMap | Descartes | 0.06 | 0.83 |
| TGFBR2 | -0.0003339 | 5072 | GTEx | DepMap | Descartes | 0.02 | 0.19 |
| IGFBP7 | -0.0003609 | 5455 | GTEx | DepMap | Descartes | 0.11 | 8.03 |
| TMEM119 | -0.0004039 | 6123 | GTEx | DepMap | Descartes | 0.02 | 0.41 |
| THY1 | -0.0004267 | 6463 | GTEx | DepMap | Descartes | 0.13 | 3.66 |
| TGFBR1 | -0.0004705 | 7083 | GTEx | DepMap | Descartes | 0.13 | 2.25 |
| TPM2 | -0.0004871 | 7322 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CNN2 | -0.0005118 | 7677 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TGFB1 | -0.0006228 | 8975 | GTEx | DepMap | Descartes | 0.02 | 0.41 |
| TPM1 | -0.0006597 | 9349 | GTEx | DepMap | Descartes | 0.89 | 21.09 |
| VCAN | -0.0007088 | 9766 | GTEx | DepMap | Descartes | 0.26 | 3.95 |
| CNN3 | -0.0008428 | 10626 | GTEx | DepMap | Descartes | 0.38 | 17.20 |
| RGS5 | -0.0008433 | 10631 | GTEx | DepMap | Descartes | 6.68 | 109.55 |
Mesenchymal Fig 1D (Olsen)
Selected mesenchymal marker genes shown in Fig. 1D of Olsen et al. https://www.biorxiv.org/content/10.1101/2020.05.04.077057v1 - these are highly expressed in their mesenchymal cluster on their UMAP.:
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.72e-03
Mean rank of genes in gene set: 3483.85
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| MGP | 0.0000192 | 415 | GTEx | DepMap | Descartes | 0.26 | 15.22 |
| PRRX1 | -0.0001100 | 1807 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| BGN | -0.0001520 | 2426 | GTEx | DepMap | Descartes | 0.02 | 0.89 |
| LUM | -0.0001580 | 2511 | GTEx | DepMap | Descartes | 0.02 | 1.43 |
| COL6A2 | -0.0001582 | 2516 | GTEx | DepMap | Descartes | 0.21 | 7.01 |
| COL1A1 | -0.0001661 | 2633 | GTEx | DepMap | Descartes | 0.15 | 2.48 |
| DCN | -0.0001710 | 2719 | GTEx | DepMap | Descartes | 0.11 | 1.53 |
| COL3A1 | -0.0002008 | 3125 | GTEx | DepMap | Descartes | 0.17 | 3.05 |
| COL1A2 | -0.0002031 | 3151 | GTEx | DepMap | Descartes | 0.17 | 2.79 |
| LEPR | -0.0002035 | 3153 | GTEx | DepMap | Descartes | 0.02 | 0.14 |
| SPARC | -0.0003219 | 4903 | GTEx | DepMap | Descartes | 0.11 | 3.90 |
| PDGFRA | -0.0004159 | 6299 | GTEx | DepMap | Descartes | 0.04 | 0.57 |
| CALD1 | -0.0006919 | 9632 | GTEx | DepMap | Descartes | 0.28 | 8.67 |
Below shows ranks on this GEP for literature curated gene lists for large gene sets
These include those reported as mesenchymal/adrenergic by Van Groningen et al.
High ranks indicate this gene is a driver of this GEP (note these results are not ordered).
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
VanGroningen Adrenergic Genes
Adrenergic marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 9289.65
Median rank of genes in gene set: 10166
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| UCP2 | 0.0003521 | 134 | GTEx | DepMap | Descartes | 0.64 | 30.17 |
| HK2 | 0.0003041 | 148 | GTEx | DepMap | Descartes | 0.06 | 1.69 |
| MYRIP | 0.0000283 | 400 | GTEx | DepMap | Descartes | 0.02 | 0.75 |
| SHC3 | -0.0000532 | 1014 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| BEND4 | -0.0000764 | 1327 | GTEx | DepMap | Descartes | 0.04 | 0.53 |
| STRA6 | -0.0000956 | 1603 | GTEx | DepMap | Descartes | 0.02 | 0.58 |
| GLDC | -0.0001345 | 2177 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| DIABLO | -0.0001535 | 2447 | GTEx | DepMap | Descartes | 0.17 | 8.72 |
| CACNA2D2 | -0.0001710 | 2720 | GTEx | DepMap | Descartes | 0.06 | 0.75 |
| MXI1 | -0.0001743 | 2767 | GTEx | DepMap | Descartes | 0.45 | 19.64 |
| RRM2 | -0.0001897 | 2961 | GTEx | DepMap | Descartes | 0.13 | 2.12 |
| NGRN | -0.0001923 | 3005 | GTEx | DepMap | Descartes | 0.26 | 7.89 |
| RNF144A | -0.0001931 | 3022 | GTEx | DepMap | Descartes | 0.17 | 2.94 |
| KIF15 | -0.0002115 | 3253 | GTEx | DepMap | Descartes | 0.06 | 0.95 |
| HS6ST2 | -0.0002145 | 3300 | GTEx | DepMap | Descartes | 0.04 | 0.62 |
| RNFT2 | -0.0002164 | 3318 | GTEx | DepMap | Descartes | 0.32 | 7.83 |
| PDK1 | -0.0002225 | 3404 | GTEx | DepMap | Descartes | 0.06 | 0.31 |
| TIAM1 | -0.0002274 | 3481 | GTEx | DepMap | Descartes | 0.04 | 1.66 |
| FOXM1 | -0.0002328 | 3557 | GTEx | DepMap | Descartes | 0.23 | 6.17 |
| CDCA5 | -0.0002447 | 3742 | GTEx | DepMap | Descartes | 0.19 | 6.60 |
| POPDC3 | -0.0002572 | 3926 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TBPL1 | -0.0002574 | 3929 | GTEx | DepMap | Descartes | 0.57 | 18.33 |
| GRB10 | -0.0002603 | 3980 | GTEx | DepMap | Descartes | 0.06 | 1.31 |
| RET | -0.0002669 | 4067 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ESRRG | -0.0002745 | 4183 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CDKN3 | -0.0002839 | 4335 | GTEx | DepMap | Descartes | 0.38 | 32.79 |
| NANOS1 | -0.0002888 | 4408 | GTEx | DepMap | Descartes | 0.09 | 1.96 |
| FAM167A | -0.0002916 | 4454 | GTEx | DepMap | Descartes | 0.02 | 0.27 |
| MCM2 | -0.0002960 | 4533 | GTEx | DepMap | Descartes | 0.19 | 4.21 |
| EML6 | -0.0002984 | 4565 | GTEx | DepMap | Descartes | 0.02 | 0.11 |
VanGroningen Mesenchymal Genes
Mesenchymal marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.45e-01
Mean rank of genes in gene set: 6094.4
Median rank of genes in gene set: 5971
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| MICAL2 | 0.0007364 | 84 | GTEx | DepMap | Descartes | 0.06 | 1.38 |
| CREG1 | 0.0003906 | 127 | GTEx | DepMap | Descartes | 0.43 | 21.16 |
| CYBRD1 | 0.0003504 | 135 | GTEx | DepMap | Descartes | 0.15 | 4.61 |
| ARL4A | 0.0003373 | 139 | GTEx | DepMap | Descartes | 0.38 | 10.61 |
| HIST1H2BK | 0.0001774 | 206 | GTEx | DepMap | Descartes | 0.02 | NA |
| GDF15 | 0.0001061 | 277 | GTEx | DepMap | Descartes | 0.02 | 1.42 |
| PXDC1 | 0.0001045 | 280 | GTEx | DepMap | Descartes | 0.02 | 0.58 |
| GAS1 | 0.0000456 | 364 | GTEx | DepMap | Descartes | 0.38 | 11.98 |
| POSTN | 0.0000366 | 384 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TRAM2 | 0.0000282 | 401 | GTEx | DepMap | Descartes | 0.02 | 0.32 |
| MGP | 0.0000192 | 415 | GTEx | DepMap | Descartes | 0.26 | 15.22 |
| F2RL2 | 0.0000125 | 435 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PROM1 | -0.0000116 | 510 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TNC | -0.0000163 | 557 | GTEx | DepMap | Descartes | 0.04 | 1.27 |
| SOSTDC1 | -0.0000241 | 653 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| GJA1 | -0.0000380 | 833 | GTEx | DepMap | Descartes | 0.09 | 2.72 |
| DLX2 | -0.0000386 | 843 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CPS1 | -0.0000482 | 954 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CAPN6 | -0.0000483 | 955 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| EPHA3 | -0.0000511 | 981 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CILP | -0.0000514 | 986 | GTEx | DepMap | Descartes | 0.02 | 0.37 |
| PRDM6 | -0.0000621 | 1129 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| BMP5 | -0.0000664 | 1192 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| DLX1 | -0.0000698 | 1247 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TNS1 | -0.0000746 | 1307 | GTEx | DepMap | Descartes | 0.04 | 0.29 |
| TFE3 | -0.0000788 | 1360 | GTEx | DepMap | Descartes | 0.11 | 3.96 |
| MEOX2 | -0.0000800 | 1378 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ELK3 | -0.0000922 | 1546 | GTEx | DepMap | Descartes | 0.04 | 0.68 |
| SYDE1 | -0.0000969 | 1622 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| AJUBA | -0.0000982 | 1632 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
Descartes adrenocortical markers
Top 50 marker genes of adrenocortical cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/adrenocortical/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 6.29e-01
Mean rank of genes in gene set: 6472.83
Median rank of genes in gene set: 6635
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| FREM2 | -0.0000601 | 1101 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SGCZ | -0.0000815 | 1397 | GTEx | DepMap | Descartes | 0.04 | 1.29 |
| STAR | -0.0000949 | 1594 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| DHCR24 | -0.0001136 | 1872 | GTEx | DepMap | Descartes | 0.11 | 1.43 |
| PAPSS2 | -0.0001424 | 2290 | GTEx | DepMap | Descartes | 0.02 | 0.27 |
| INHA | -0.0001717 | 2728 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| BAIAP2L1 | -0.0001750 | 2776 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| APOC1 | -0.0002817 | 4294 | GTEx | DepMap | Descartes | 1.32 | 151.21 |
| FDXR | -0.0003042 | 4648 | GTEx | DepMap | Descartes | 0.09 | 3.53 |
| NPC1 | -0.0003154 | 4806 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| GRAMD1B | -0.0003330 | 5062 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SCARB1 | -0.0003350 | 5086 | GTEx | DepMap | Descartes | 0.06 | 0.69 |
| SH3PXD2B | -0.0003558 | 5385 | GTEx | DepMap | Descartes | 0.02 | 0.51 |
| FRMD5 | -0.0003574 | 5397 | GTEx | DepMap | Descartes | 0.02 | 0.27 |
| ERN1 | -0.0003764 | 5703 | GTEx | DepMap | Descartes | 0.04 | 0.49 |
| SCAP | -0.0003941 | 5978 | GTEx | DepMap | Descartes | 0.06 | 2.66 |
| CYB5B | -0.0004153 | 6287 | GTEx | DepMap | Descartes | 0.49 | 14.36 |
| CLU | -0.0004280 | 6483 | GTEx | DepMap | Descartes | 0.91 | 31.70 |
| PDE10A | -0.0004485 | 6787 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| DHCR7 | -0.0004537 | 6864 | GTEx | DepMap | Descartes | 0.11 | 3.28 |
| LDLR | -0.0004703 | 7078 | GTEx | DepMap | Descartes | 0.04 | 0.43 |
| SLC16A9 | -0.0004734 | 7140 | GTEx | DepMap | Descartes | 0.02 | 0.28 |
| HMGCR | -0.0004964 | 7448 | GTEx | DepMap | Descartes | 0.21 | 3.89 |
| SLC1A2 | -0.0005362 | 8007 | GTEx | DepMap | Descartes | 0.11 | 1.31 |
| DNER | -0.0005372 | 8020 | GTEx | DepMap | Descartes | 0.13 | 4.65 |
| POR | -0.0005908 | 8607 | GTEx | DepMap | Descartes | 0.04 | 2.40 |
| PEG3 | -0.0006352 | 9108 | GTEx | DepMap | Descartes | 0.23 | NA |
| TM7SF2 | -0.0006495 | 9241 | GTEx | DepMap | Descartes | 0.30 | 18.05 |
| HMGCS1 | -0.0006530 | 9283 | GTEx | DepMap | Descartes | 0.30 | 3.21 |
| MSMO1 | -0.0006600 | 9353 | GTEx | DepMap | Descartes | 0.15 | 10.01 |
Descartes chromaffin markers
Top 50 marker genes of chromaffin cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/chromaffin/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 9314.76
Median rank of genes in gene set: 9992
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| SLC44A5 | -0.0001998 | 3110 | GTEx | DepMap | Descartes | 0.02 | 0.35 |
| EPHA6 | -0.0002557 | 3906 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TMEM132C | -0.0002744 | 4182 | GTEx | DepMap | Descartes | 0.09 | 1.17 |
| RPH3A | -0.0003187 | 4856 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ALK | -0.0003194 | 4864 | GTEx | DepMap | Descartes | 0.06 | 0.75 |
| ANKFN1 | -0.0003197 | 4870 | GTEx | DepMap | Descartes | 0.02 | 0.25 |
| CNKSR2 | -0.0003263 | 4968 | GTEx | DepMap | Descartes | 0.09 | 1.42 |
| GREM1 | -0.0003265 | 4971 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| KCNB2 | -0.0004688 | 7060 | GTEx | DepMap | Descartes | 0.06 | 4.39 |
| FAT3 | -0.0004901 | 7366 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| RYR2 | -0.0004969 | 7457 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SLC6A2 | -0.0004981 | 7477 | GTEx | DepMap | Descartes | 0.17 | 6.15 |
| EYA1 | -0.0005136 | 7704 | GTEx | DepMap | Descartes | 0.04 | 0.73 |
| EYA4 | -0.0005559 | 8218 | GTEx | DepMap | Descartes | 0.02 | 0.68 |
| SYNPO2 | -0.0005942 | 8646 | GTEx | DepMap | Descartes | 0.13 | 1.75 |
| HS3ST5 | -0.0006052 | 8767 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PTCHD1 | -0.0006361 | 9116 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PLXNA4 | -0.0006390 | 9145 | GTEx | DepMap | Descartes | 0.02 | 0.08 |
| GAL | -0.0006783 | 9513 | GTEx | DepMap | Descartes | 1.28 | 182.63 |
| RGMB | -0.0007071 | 9749 | GTEx | DepMap | Descartes | 0.30 | 5.19 |
| MAB21L2 | -0.0007405 | 9992 | GTEx | DepMap | Descartes | 0.57 | 28.39 |
| TMEFF2 | -0.0007956 | 10387 | GTEx | DepMap | Descartes | 0.11 | 3.95 |
| MAB21L1 | -0.0008823 | 10844 | GTEx | DepMap | Descartes | 0.57 | 24.16 |
| RBFOX1 | -0.0009401 | 11095 | GTEx | DepMap | Descartes | 0.11 | 3.67 |
| NTRK1 | -0.0009482 | 11131 | GTEx | DepMap | Descartes | 0.09 | 3.60 |
| IL7 | -0.0009740 | 11215 | GTEx | DepMap | Descartes | 0.53 | 36.08 |
| MARCH11 | -0.0010403 | 11448 | GTEx | DepMap | Descartes | 1.09 | NA |
| REEP1 | -0.0010432 | 11461 | GTEx | DepMap | Descartes | 0.09 | 2.46 |
| CNTFR | -0.0011386 | 11687 | GTEx | DepMap | Descartes | 0.21 | 15.19 |
| ISL1 | -0.0011483 | 11712 | GTEx | DepMap | Descartes | 1.26 | 83.76 |
Descartes Vascular_endothelial markers
Top 50 marker genes of Vascular_endothelial cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/vascular_endothelial/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 3.11e-08
Mean rank of genes in gene set: 3098.92
Median rank of genes in gene set: 2536
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CDH13 | 0.0002088 | 188 | GTEx | DepMap | Descartes | 0.09 | 1.75 |
| ESM1 | 0.0001580 | 225 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CRHBP | 0.0000332 | 391 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MYRIP | 0.0000283 | 400 | GTEx | DepMap | Descartes | 0.02 | 0.75 |
| PLVAP | 0.0000119 | 437 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TIE1 | -0.0000347 | 799 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| KDR | -0.0000668 | 1198 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SLCO2A1 | -0.0000806 | 1384 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| NR5A2 | -0.0000836 | 1428 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| FLT4 | -0.0000843 | 1441 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| NPR1 | -0.0000852 | 1457 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| GALNT15 | -0.0001090 | 1787 | GTEx | DepMap | Descartes | 0.00 | NA |
| PTPRB | -0.0001112 | 1835 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CALCRL | -0.0001194 | 1951 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| BTNL9 | -0.0001280 | 2088 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SHE | -0.0001299 | 2113 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TEK | -0.0001315 | 2137 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| KANK3 | -0.0001383 | 2225 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MMRN2 | -0.0001561 | 2488 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ROBO4 | -0.0001624 | 2584 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CLDN5 | -0.0001662 | 2638 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CDH5 | -0.0001712 | 2721 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| RASIP1 | -0.0001885 | 2943 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| EHD3 | -0.0002094 | 3228 | GTEx | DepMap | Descartes | 0.04 | 0.55 |
| CEACAM1 | -0.0002188 | 3364 | GTEx | DepMap | Descartes | 0.02 | 1.63 |
| F8 | -0.0002225 | 3402 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PODXL | -0.0002320 | 3543 | GTEx | DepMap | Descartes | 0.06 | 1.05 |
| TMEM88 | -0.0002466 | 3775 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| IRX3 | -0.0002737 | 4173 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ID1 | -0.0002822 | 4299 | GTEx | DepMap | Descartes | 0.06 | 9.70 |
Descartes stromal markers
Top 50 marker genes of stromal cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/stromal/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 5.70e-11
Mean rank of genes in gene set: 2760.14
Median rank of genes in gene set: 2156.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| RSPO3 | 0.0001221 | 258 | GTEx | DepMap | Descartes | 0.04 | NA |
| POSTN | 0.0000366 | 384 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MGP | 0.0000192 | 415 | GTEx | DepMap | Descartes | 0.26 | 15.22 |
| C7 | 0.0000074 | 448 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ABCC9 | -0.0000258 | 678 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| IGFBP3 | -0.0000319 | 773 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| LAMC3 | -0.0000387 | 844 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ITGA11 | -0.0000449 | 910 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SFRP2 | -0.0000797 | 1372 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SCARA5 | -0.0000938 | 1570 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| LOX | -0.0000939 | 1575 | GTEx | DepMap | Descartes | 0.02 | 1.12 |
| DKK2 | -0.0000990 | 1643 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| EDNRA | -0.0001077 | 1766 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ADAMTSL3 | -0.0001088 | 1786 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| COL6A3 | -0.0001091 | 1790 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PRRX1 | -0.0001100 | 1807 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| COL27A1 | -0.0001101 | 1813 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| GLI2 | -0.0001106 | 1822 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CCDC80 | -0.0001199 | 1958 | GTEx | DepMap | Descartes | 0.32 | 2.53 |
| GAS2 | -0.0001213 | 1990 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PAMR1 | -0.0001263 | 2069 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| LRRC17 | -0.0001314 | 2134 | GTEx | DepMap | Descartes | 0.02 | 0.74 |
| ISLR | -0.0001347 | 2179 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ABCA6 | -0.0001454 | 2336 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| BICC1 | -0.0001518 | 2425 | GTEx | DepMap | Descartes | 0.04 | 0.73 |
| OGN | -0.0001564 | 2493 | GTEx | DepMap | Descartes | 0.02 | 0.61 |
| LUM | -0.0001580 | 2511 | GTEx | DepMap | Descartes | 0.02 | 1.43 |
| PCDH18 | -0.0001632 | 2591 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| COL1A1 | -0.0001661 | 2633 | GTEx | DepMap | Descartes | 0.15 | 2.48 |
| DCN | -0.0001710 | 2719 | GTEx | DepMap | Descartes | 0.11 | 1.53 |
Descartes sympathoblasts markers
Top 50 marker genes of sympathoblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/sympathoblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.25e-01
Mean rank of genes in gene set: 5599.39
Median rank of genes in gene set: 4701
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| DGKK | -0.0000526 | 1001 | GTEx | DepMap | Descartes | 0.02 | 0.51 |
| KSR2 | -0.0000635 | 1153 | GTEx | DepMap | Descartes | 0.06 | 0.51 |
| PENK | -0.0000813 | 1392 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ST18 | -0.0000956 | 1602 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PCSK2 | -0.0001637 | 2603 | GTEx | DepMap | Descartes | 0.02 | 0.29 |
| SORCS3 | -0.0001775 | 2815 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CNTN3 | -0.0001778 | 2820 | GTEx | DepMap | Descartes | 0.02 | 0.22 |
| LAMA3 | -0.0001865 | 2916 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SLC35F3 | -0.0001895 | 2955 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TENM1 | -0.0001922 | 3000 | GTEx | DepMap | Descartes | 0.06 | NA |
| TBX20 | -0.0001996 | 3108 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| GRM7 | -0.0002104 | 3242 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TIAM1 | -0.0002274 | 3481 | GTEx | DepMap | Descartes | 0.04 | 1.66 |
| SLC24A2 | -0.0002305 | 3525 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CDH12 | -0.0002442 | 3734 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CDH18 | -0.0002493 | 3811 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SLC18A1 | -0.0002941 | 4493 | GTEx | DepMap | Descartes | 0.11 | 4.91 |
| EML6 | -0.0002984 | 4565 | GTEx | DepMap | Descartes | 0.02 | 0.11 |
| GALNTL6 | -0.0002999 | 4589 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CCSER1 | -0.0003161 | 4813 | GTEx | DepMap | Descartes | 0.00 | NA |
| AGBL4 | -0.0003199 | 4874 | GTEx | DepMap | Descartes | 0.04 | 0.57 |
| MGAT4C | -0.0003810 | 5769 | GTEx | DepMap | Descartes | 0.11 | 0.32 |
| FGF14 | -0.0003959 | 6015 | GTEx | DepMap | Descartes | 0.13 | 1.73 |
| NTNG1 | -0.0004387 | 6641 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SPOCK3 | -0.0004395 | 6653 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| GRID2 | -0.0004492 | 6804 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PACRG | -0.0004666 | 7029 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| FAM155A | -0.0005196 | 7801 | GTEx | DepMap | Descartes | 0.11 | 1.80 |
| UNC80 | -0.0005518 | 8184 | GTEx | DepMap | Descartes | 0.04 | 0.19 |
| CHGA | -0.0005692 | 8363 | GTEx | DepMap | Descartes | 2.17 | 113.77 |
Descartes erythroblasts markers
Top 50 marker genes of erythroblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/erythroblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.66e-12
Mean rank of genes in gene set: 1640.83
Median rank of genes in gene set: 274
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ALAS2 | 0.0642696 | 1 | GTEx | DepMap | Descartes | 10.13 | 555.57 |
| SELENBP1 | 0.0274233 | 2 | GTEx | DepMap | Descartes | 1.11 | 34.19 |
| SLC25A37 | 0.0265062 | 3 | GTEx | DepMap | Descartes | 6.62 | 151.57 |
| SLC4A1 | 0.0203923 | 5 | GTEx | DepMap | Descartes | 0.57 | 10.08 |
| SNCA | 0.0119594 | 9 | GTEx | DepMap | Descartes | 4.38 | 137.78 |
| RHD | 0.0081725 | 13 | GTEx | DepMap | Descartes | 0.09 | 3.24 |
| GYPC | 0.0066174 | 17 | GTEx | DepMap | Descartes | 4.53 | 296.75 |
| BLVRB | 0.0060679 | 19 | GTEx | DepMap | Descartes | 2.87 | 205.16 |
| FECH | 0.0018103 | 51 | GTEx | DepMap | Descartes | 0.21 | 3.63 |
| CAT | 0.0017772 | 52 | GTEx | DepMap | Descartes | 0.36 | 12.36 |
| ANK1 | 0.0011899 | 61 | GTEx | DepMap | Descartes | 0.32 | 3.66 |
| MICAL2 | 0.0007364 | 84 | GTEx | DepMap | Descartes | 0.06 | 1.38 |
| TMCC2 | 0.0005207 | 107 | GTEx | DepMap | Descartes | 0.15 | 3.41 |
| MARCH3 | 0.0001937 | 195 | GTEx | DepMap | Descartes | 0.04 | NA |
| GCLC | 0.0001078 | 274 | GTEx | DepMap | Descartes | 0.13 | 1.97 |
| CPOX | 0.0000945 | 292 | GTEx | DepMap | Descartes | 0.09 | 2.18 |
| EPB41 | 0.0000510 | 356 | GTEx | DepMap | Descartes | 0.32 | 4.78 |
| SLC25A21 | -0.0000239 | 649 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SPECC1 | -0.0000311 | 761 | GTEx | DepMap | Descartes | 0.09 | 1.54 |
| TFR2 | -0.0000604 | 1106 | GTEx | DepMap | Descartes | 0.26 | 5.27 |
| RGS6 | -0.0000890 | 1501 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| XPO7 | -0.0001605 | 2554 | GTEx | DepMap | Descartes | 0.06 | 1.52 |
| TSPAN5 | -0.0001807 | 2852 | GTEx | DepMap | Descartes | 0.45 | 11.52 |
| SOX6 | -0.0002589 | 3958 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ABCB10 | -0.0002954 | 4519 | GTEx | DepMap | Descartes | 0.02 | 0.30 |
| TRAK2 | -0.0003423 | 5202 | GTEx | DepMap | Descartes | 0.13 | 1.79 |
| SPTB | -0.0003710 | 5614 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| RAPGEF2 | -0.0005746 | 8429 | GTEx | DepMap | Descartes | 0.04 | 1.32 |
| DENND4A | -0.0006155 | 8898 | GTEx | DepMap | Descartes | 0.09 | 0.68 |
| NA | NA | NA | NA | NA | NA | NA | NA |
Descartes myeloid markers
Top 50 marker genes of myeloid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/myeloid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 5.98e-02
Mean rank of genes in gene set: 5361.5
Median rank of genes in gene set: 5076
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| SFMBT2 | 0.0000502 | 357 | GTEx | DepMap | Descartes | 0.04 | 0.40 |
| FGD2 | -0.0001656 | 2629 | GTEx | DepMap | Descartes | 0.02 | 0.20 |
| MERTK | -0.0001752 | 2781 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CPVL | -0.0001869 | 2923 | GTEx | DepMap | Descartes | 0.02 | 0.52 |
| ATP8B4 | -0.0002098 | 3233 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| FGL2 | -0.0002245 | 3434 | GTEx | DepMap | Descartes | 0.02 | 0.42 |
| MS4A4A | -0.0002285 | 3491 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SLC1A3 | -0.0002288 | 3498 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| HRH1 | -0.0002301 | 3516 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PTPRE | -0.0002454 | 3751 | GTEx | DepMap | Descartes | 0.04 | 0.72 |
| SLC9A9 | -0.0002488 | 3801 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CD163 | -0.0002693 | 4109 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MARCH1 | -0.0002824 | 4303 | GTEx | DepMap | Descartes | 0.02 | NA |
| CYBB | -0.0002825 | 4308 | GTEx | DepMap | Descartes | 0.02 | 0.44 |
| SLCO2B1 | -0.0002848 | 4347 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CSF1R | -0.0002864 | 4370 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| RGL1 | -0.0002936 | 4484 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ADAP2 | -0.0003225 | 4910 | GTEx | DepMap | Descartes | 0.02 | 1.33 |
| AXL | -0.0003328 | 5061 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ITPR2 | -0.0003353 | 5091 | GTEx | DepMap | Descartes | 0.06 | 0.69 |
| SPP1 | -0.0003415 | 5189 | GTEx | DepMap | Descartes | 0.91 | 59.39 |
| ABCA1 | -0.0003493 | 5306 | GTEx | DepMap | Descartes | 0.06 | 0.85 |
| HCK | -0.0003516 | 5325 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MSR1 | -0.0003538 | 5354 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CD14 | -0.0003577 | 5404 | GTEx | DepMap | Descartes | 0.02 | 1.70 |
| LGMN | -0.0003899 | 5912 | GTEx | DepMap | Descartes | 0.06 | 2.84 |
| TGFBI | -0.0004148 | 6275 | GTEx | DepMap | Descartes | 0.02 | 0.83 |
| CTSC | -0.0004391 | 6646 | GTEx | DepMap | Descartes | 0.09 | 1.01 |
| CTSS | -0.0004495 | 6808 | GTEx | DepMap | Descartes | 0.04 | 0.89 |
| CTSB | -0.0004502 | 6821 | GTEx | DepMap | Descartes | 0.49 | 14.28 |
Descartes Schwann markers
Top 50 marker genes of Schwann cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/schwann/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.19e-01
Mean rank of genes in gene set: 5852.3
Median rank of genes in gene set: 4920.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| IL1RAPL2 | -0.0000116 | 509 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| HMGA2 | -0.0000637 | 1155 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PTPRZ1 | -0.0000965 | 1615 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ADAMTS5 | -0.0001241 | 2034 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MDGA2 | -0.0001293 | 2104 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| IL1RAPL1 | -0.0001414 | 2277 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MPZ | -0.0001606 | 2556 | GTEx | DepMap | Descartes | 0.04 | 4.03 |
| OLFML2A | -0.0001679 | 2667 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TRPM3 | -0.0001773 | 2813 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| STARD13 | -0.0001787 | 2830 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| GRIK3 | -0.0001842 | 2893 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ERBB4 | -0.0001921 | 2997 | GTEx | DepMap | Descartes | 0.02 | 0.17 |
| EDNRB | -0.0002048 | 3166 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| NRXN3 | -0.0002219 | 3399 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ERBB3 | -0.0002270 | 3474 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| COL25A1 | -0.0002365 | 3609 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PLP1 | -0.0002413 | 3688 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| COL5A2 | -0.0002547 | 3892 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| COL18A1 | -0.0002548 | 3893 | GTEx | DepMap | Descartes | 0.11 | 1.55 |
| XKR4 | -0.0002855 | 4354 | GTEx | DepMap | Descartes | 0.06 | 0.39 |
| PLCE1 | -0.0003000 | 4593 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| EGFLAM | -0.0003152 | 4802 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SLC35F1 | -0.0003315 | 5039 | GTEx | DepMap | Descartes | 0.02 | 0.76 |
| LRRTM4 | -0.0003387 | 5146 | GTEx | DepMap | Descartes | 0.02 | 0.43 |
| GAS7 | -0.0003645 | 5512 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| LAMC1 | -0.0004014 | 6090 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SORCS1 | -0.0004276 | 6479 | GTEx | DepMap | Descartes | 0.13 | 2.64 |
| SCN7A | -0.0004289 | 6493 | GTEx | DepMap | Descartes | 0.06 | 1.07 |
| KCTD12 | -0.0004784 | 7201 | GTEx | DepMap | Descartes | 0.02 | 0.34 |
| LAMA4 | -0.0004829 | 7264 | GTEx | DepMap | Descartes | 0.09 | 1.16 |
Descartes Megakaryocytes markers
Top 50 marker genes of Megakaryocytes cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/megakaryocytes/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.60e-01
Mean rank of genes in gene set: 5739.91
Median rank of genes in gene set: 4670
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TUBB1 | 0.0022514 | 44 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| STOM | 0.0011883 | 62 | GTEx | DepMap | Descartes | 0.21 | 5.82 |
| GP1BA | -0.0000198 | 592 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ITGB3 | -0.0000617 | 1125 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MMRN1 | -0.0000771 | 1338 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SPN | -0.0001062 | 1749 | GTEx | DepMap | Descartes | 0.13 | 0.95 |
| SLC24A3 | -0.0001103 | 1816 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| P2RX1 | -0.0001146 | 1882 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TRPC6 | -0.0001216 | 1992 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ARHGAP6 | -0.0001425 | 2292 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ANGPT1 | -0.0001703 | 2708 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CD84 | -0.0001926 | 3010 | GTEx | DepMap | Descartes | 0.04 | 0.68 |
| FLI1 | -0.0002119 | 3261 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ITGA2B | -0.0002158 | 3315 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| UBASH3B | -0.0002250 | 3443 | GTEx | DepMap | Descartes | 0.04 | 0.36 |
| PLEK | -0.0002572 | 3925 | GTEx | DepMap | Descartes | 0.02 | 0.69 |
| MED12L | -0.0002652 | 4050 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| LTBP1 | -0.0002655 | 4051 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| INPP4B | -0.0002680 | 4084 | GTEx | DepMap | Descartes | 0.11 | 1.46 |
| GSN | -0.0002767 | 4212 | GTEx | DepMap | Descartes | 0.04 | 0.98 |
| MYLK | -0.0002805 | 4268 | GTEx | DepMap | Descartes | 0.06 | 0.63 |
| THBS1 | -0.0002878 | 4398 | GTEx | DepMap | Descartes | 0.06 | 0.81 |
| RAB27B | -0.0003050 | 4670 | GTEx | DepMap | Descartes | 0.04 | 1.05 |
| MCTP1 | -0.0003052 | 4674 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PSTPIP2 | -0.0003118 | 4753 | GTEx | DepMap | Descartes | 0.02 | 0.38 |
| PDE3A | -0.0003145 | 4791 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MYH9 | -0.0004176 | 6327 | GTEx | DepMap | Descartes | 0.19 | 2.72 |
| STON2 | -0.0004386 | 6638 | GTEx | DepMap | Descartes | 0.02 | 0.25 |
| DOK6 | -0.0005052 | 7575 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TLN1 | -0.0005160 | 7740 | GTEx | DepMap | Descartes | 0.15 | 1.09 |
Descartes Lyphoid markers
Top 50 marker genes of Lyphoid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/lymphoid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 8120.62
Median rank of genes in gene set: 7928
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| SP100 | -0.0000915 | 1532 | GTEx | DepMap | Descartes | 0.19 | 3.27 |
| PLEKHA2 | -0.0001098 | 1804 | GTEx | DepMap | Descartes | 0.09 | 1.65 |
| ITPKB | -0.0002059 | 3179 | GTEx | DepMap | Descartes | 0.02 | 0.29 |
| SAMD3 | -0.0002455 | 3754 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| MCTP2 | -0.0002791 | 4244 | GTEx | DepMap | Descartes | 0.02 | 1.02 |
| CCL5 | -0.0003485 | 5298 | GTEx | DepMap | Descartes | 0.13 | 6.36 |
| LEF1 | -0.0003729 | 5645 | GTEx | DepMap | Descartes | 0.09 | 3.14 |
| ETS1 | -0.0003875 | 5875 | GTEx | DepMap | Descartes | 0.06 | 1.89 |
| BACH2 | -0.0003919 | 5945 | GTEx | DepMap | Descartes | 0.04 | 0.29 |
| RAP1GAP2 | -0.0003955 | 6003 | GTEx | DepMap | Descartes | 0.17 | 2.24 |
| SKAP1 | -0.0004116 | 6234 | GTEx | DepMap | Descartes | 0.02 | 2.61 |
| PDE3B | -0.0004455 | 6743 | GTEx | DepMap | Descartes | 0.04 | 1.14 |
| SCML4 | -0.0004643 | 6999 | GTEx | DepMap | Descartes | 0.06 | 1.56 |
| PITPNC1 | -0.0004681 | 7045 | GTEx | DepMap | Descartes | 0.06 | 1.48 |
| WIPF1 | -0.0004773 | 7192 | GTEx | DepMap | Descartes | 0.09 | 1.37 |
| DOCK10 | -0.0004805 | 7240 | GTEx | DepMap | Descartes | 0.02 | 0.21 |
| PRKCH | -0.0004857 | 7307 | GTEx | DepMap | Descartes | 0.02 | 0.32 |
| IKZF1 | -0.0004876 | 7328 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| LCP1 | -0.0004947 | 7427 | GTEx | DepMap | Descartes | 0.11 | 3.62 |
| ARID5B | -0.0005170 | 7756 | GTEx | DepMap | Descartes | 0.09 | 1.94 |
| ARHGAP15 | -0.0005299 | 7915 | GTEx | DepMap | Descartes | 0.02 | 1.28 |
| RCSD1 | -0.0005313 | 7941 | GTEx | DepMap | Descartes | 0.02 | 0.20 |
| MBNL1 | -0.0005349 | 7993 | GTEx | DepMap | Descartes | 0.09 | 1.63 |
| ABLIM1 | -0.0005358 | 8003 | GTEx | DepMap | Descartes | 0.11 | 1.27 |
| PTPRC | -0.0006096 | 8812 | GTEx | DepMap | Descartes | 0.06 | 1.08 |
| TOX | -0.0006283 | 9025 | GTEx | DepMap | Descartes | 0.06 | 2.79 |
| ANKRD44 | -0.0006804 | 9536 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CCND3 | -0.0006822 | 9552 | GTEx | DepMap | Descartes | 0.19 | 8.25 |
| MSN | -0.0006941 | 9655 | GTEx | DepMap | Descartes | 0.06 | 0.79 |
| SORL1 | -0.0007319 | 9934 | GTEx | DepMap | Descartes | 0.04 | 0.47 |
Below shows the ranks on this GEP for immune marker gene lists curated by CellTypist (https://www.celltypist.org/encyclopedia).
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Erythroid: Early erythroid (model markers)
early erythroid cells which are committed to erythroid lineage cells:
Wilcoxon ranksum test P-value for gene set overrepresentation: 4.45e-03
Mean rank of genes in gene set: 1536.75
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| FAM178B | 0.0037072 | 31 | GTEx | DepMap | Descartes | 0.77 | 15.39 |
| PRSS57 | 0.0001323 | 250 | GTEx | DepMap | Descartes | 0.09 | 4.23 |
| CD207 | -0.0000938 | 1572 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| APOC1 | -0.0002817 | 4294 | GTEx | DepMap | Descartes | 1.32 | 151.21 |
B-cell lineage: Small pre-B cells (model markers)
non-proliferative B lymphocyte precursors derived from Pro-B cells and expressing membrane μ chains with surrogate light chains in their receptors:
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.11e-02
Mean rank of genes in gene set: 2135.5
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TLDC2 | -0.0000188 | 579 | GTEx | DepMap | Descartes | 0.00 | NA |
| AICDA | -0.0000862 | 1470 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CD207 | -0.0000938 | 1572 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| CD79B | -0.0003234 | 4921 | GTEx | DepMap | Descartes | 0.11 | 10.69 |
Fibroblasts: Fibroblasts (model markers)
the most common cells of connective tissues which synthesize the extracellular matrix and collagen to maintain tissue homeostasis:
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.12e-02
Mean rank of genes in gene set: 3520
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| OLFML1 | -0.0000253 | 668 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SMOC2 | -0.0000743 | 1304 | GTEx | DepMap | Descartes | 0.02 | 0.66 |
| F10 | -0.0000873 | 1481 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ANGPTL1 | -0.0001017 | 1680 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PRRX1 | -0.0001100 | 1807 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| EBF2 | -0.0001461 | 2345 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| NTRK2 | -0.0003128 | 4769 | GTEx | DepMap | Descartes | 0.13 | 3.19 |
| PDGFRA | -0.0004159 | 6299 | GTEx | DepMap | Descartes | 0.04 | 0.57 |
| SFRP1 | -0.0010012 | 11327 | GTEx | DepMap | Descartes | 0.02 | 1.27 |