Program: 21. PDX Human #21.
Program description and justification of annotation: 21.
Submit a comment on this gene expression program’s interpretation: CLICK
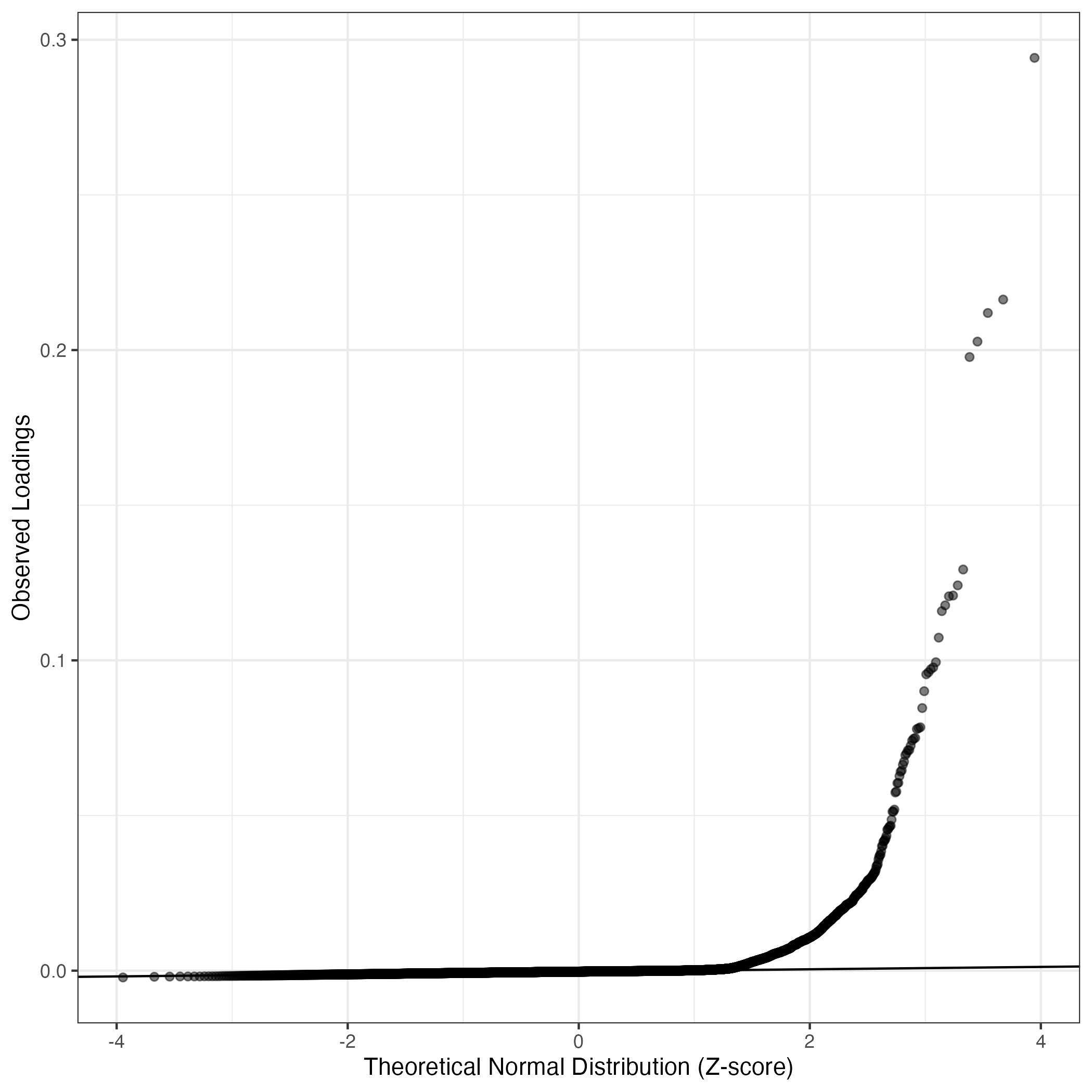
QQ-plot of gene loadings, averaged over both independent splits of the data
This plot highlights the relative contribution of each gene to the GEP
## Error in apply(allData[cellsHGreaterThan50, ], 2, mean): dim(X) must have a positive length
## Error in apply(allDataTpm[cellsHGreaterThan50, ], 2, mean): dim(X) must have a positive length
## Error in eval(expr, envir, enclos): object 'meanCountsHOver50' not found
## Error in eval(expr, envir, enclos): object 'meanTpmHOver50' not found
## Error in data.frame(Gene = geneCardsLink, Loading = dftoDisplay[, 2], : object 'countHighScoringCells' not found
## Error in rownames(dftForKable) <- rn: object 'dftForKable' not found
Top genes driving this program.
Note: Decartes website is buggy, try refreshing. Also, Decartes fetal adrenal data have been collected at specific time points (89-122 days), all possible cell types of interest may not be represented, do not overinterpret.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
## Error in kable(dftForKable[1:50, ], row.names = TRUE): object 'dftForKable' not found
## Error in eval(quote(list(...)), env): object 'countHighScoringCells' not found
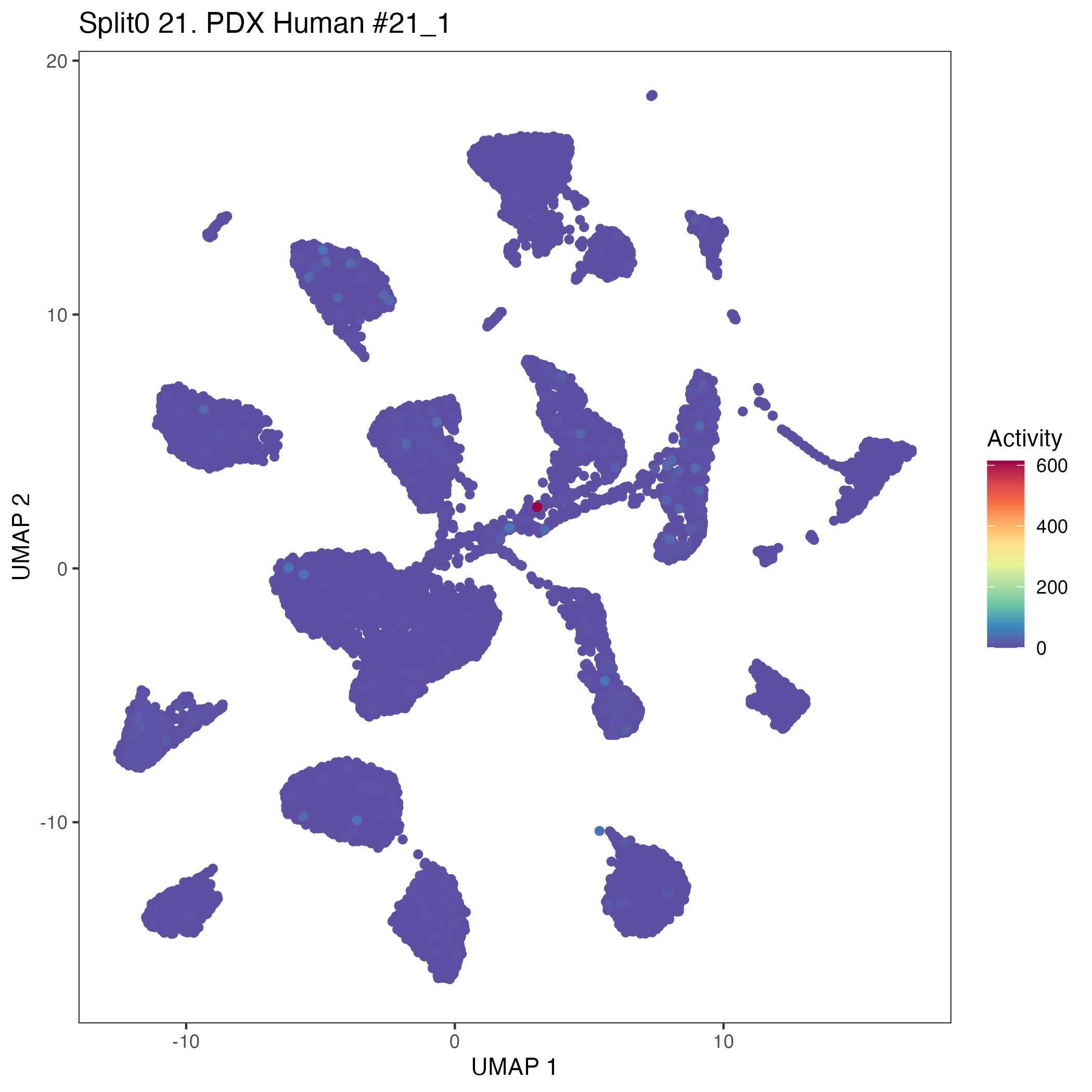
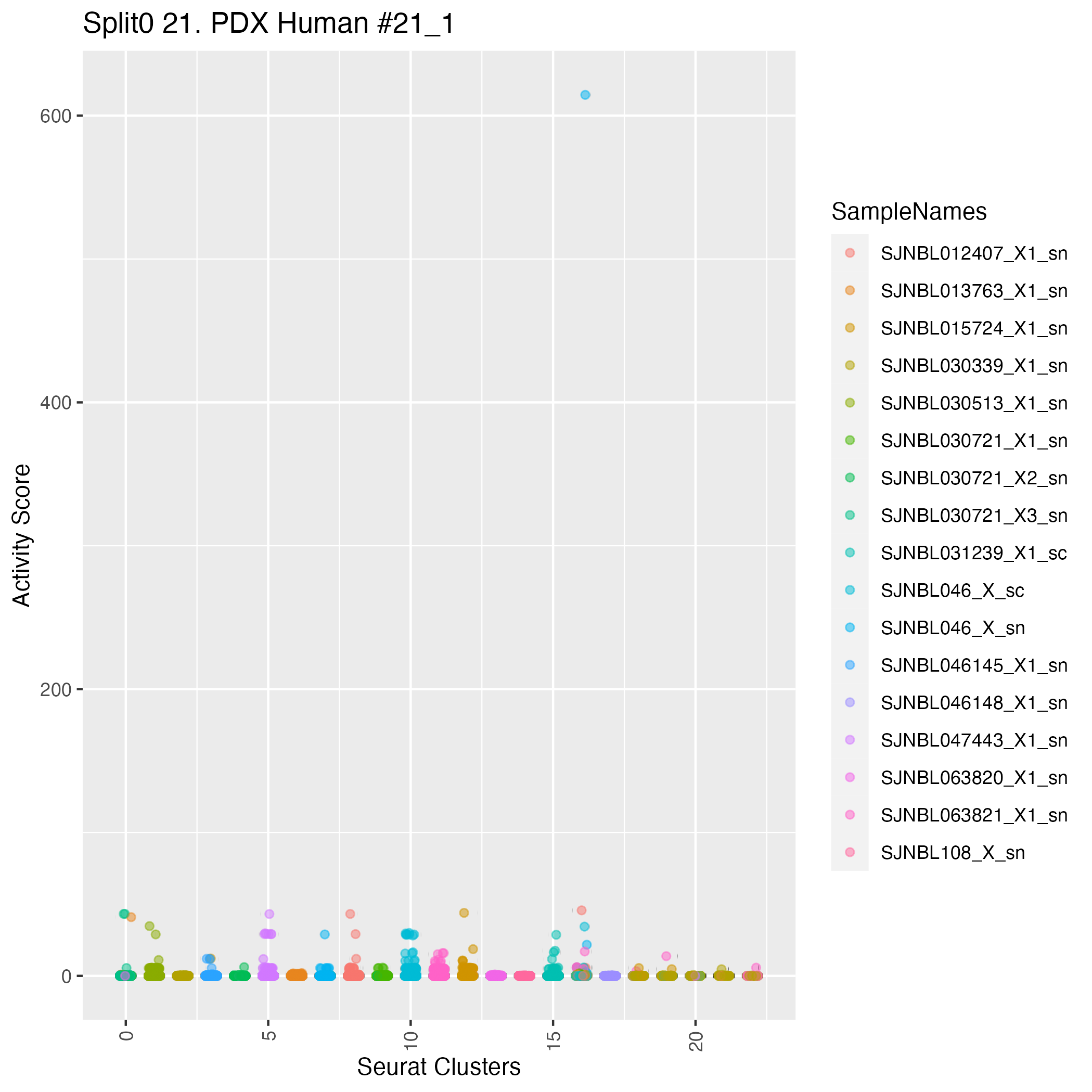
UMAP plots showing activity of gene expression program identified in community:21. PDX Human #21
Gene set Enrichments for this program, caculated from top 50 genes
mSigDB Cell Types Gene Set:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| DURANTE_ADULT_OLFACTORY_NEUROEPITHELIUM_RESPIRATORY_CILIATED_CELLS | 1.71e-17 | 37.96 | 18.83 | 1.15e-14 | 1.15e-14 | 15CRIP1, CCDC17, ODF3B, PSCA, TMEM190, SMIM22, TPPP3, FOXJ1, DNAAF1, RSPH1, MORN2, CIB1, TUBB4B, CETN2, EFCAB1 |
156 |
| DURANTE_ADULT_OLFACTORY_NEUROEPITHELIUM_UNSPECIFIED | 6.10e-09 | 54.80 | 17.80 | 2.93e-07 | 4.10e-06 | 6TPPP3, RSPH1, MORN2, CIB1, TUBB4B, CETN2 |
37 |
| DURANTE_ADULT_OLFACTORY_NEUROEPITHELIUM_RESPIRATORY_HORIZONTAL_BASAL_CELLS | 1.76e-07 | 49.45 | 14.27 | 6.57e-06 | 1.18e-04 | 5ANXA1, KRT18, AQP3, S100A11, KRT8 |
33 |
| AIZARANI_LIVER_C24_EPCAM_POS_BILE_DUCT_CELLS_3 | 4.64e-12 | 22.80 | 10.66 | 7.78e-10 | 3.11e-09 | 12S100A10, KRT18, SLPI, AQP3, TPPP3, MAL2, S100A11, S100A6, TUBB4B, TNFRSF12A, KRT8, LGALS3 |
185 |
| FAN_EMBRYONIC_CTX_BIG_GROUPS_BRAIN_IMMUNE | 2.35e-10 | 22.64 | 9.89 | 2.25e-08 | 1.58e-07 | 10ANXA1, S100A10, CRIP1, S100A4, IFITM2, S100A11, S100A6, CIB1, UCP2, B2M |
148 |
| FAN_OVARY_CL12_T_LYMPHOCYTE_NK_CELL_2 | 5.34e-10 | 20.69 | 9.06 | 4.38e-08 | 3.58e-07 | 10ANXA1, CRIP1, RGCC, S100A4, S100A11, S100A6, CIB1, TUBB4B, B2M, CYBA |
161 |
| BUSSLINGER_GASTRIC_IMMATURE_PIT_CELLS | 6.77e-10 | 20.15 | 8.83 | 4.54e-08 | 4.54e-07 | 10KRT18, CRIP1, PSCA, MAL2, S100A11, S100A6, KRT8, ITGB4, IFI27, LGALS3 |
165 |
| MENON_FETAL_KIDNEY_6_COLLECTING_DUCT_CELLS | 7.71e-08 | 23.36 | 8.63 | 3.45e-06 | 5.17e-05 | 7S100A10, KRT18, MAL2, S100A11, S100A6, KRT8, B2M |
94 |
| DESCARTES_FETAL_EYE_CORNEAL_AND_CONJUNCTIVAL_EPITHELIAL_CELLS | 1.08e-10 | 17.09 | 8.02 | 1.39e-08 | 7.24e-08 | 12ANXA1, KRT18, CRIP1, PSCA, SMIM22, SLPI, AQP3, MAL2, S100A6, KRT8, LGALS3, SLC9A3R1 |
243 |
| FAN_OVARY_CL4_T_LYMPHOCYTE_NK_CELL_1 | 1.24e-10 | 16.87 | 7.92 | 1.39e-08 | 8.33e-08 | 12ANXA1, S100A10, CRIP1, RGCC, S100A4, IFITM2, S100A11, S100A6, CIB1, TUBB4B, B2M, CYBA |
246 |
| BUSSLINGER_GASTRIC_MATURE_PIT_CELLS | 7.73e-10 | 16.55 | 7.54 | 4.72e-08 | 5.19e-07 | 11CRIP1, PSCA, SMIM22, SLPI, MAL2, S100A11, S100A6, DUOX2, KRT8, IFI27, LGALS3 |
224 |
| DESCARTES_FETAL_STOMACH_CILIATED_EPITHELIAL_CELLS | 1.85e-09 | 13.12 | 6.18 | 9.57e-08 | 1.24e-06 | 12CCDC17, ODF3B, TMEM190, TPPP3, FOXJ1, DNAAF1, RSPH1, MORN2, TTC29, SPEF1, EFCAB1, DNAH12 |
313 |
| DURANTE_ADULT_OLFACTORY_NEUROEPITHELIUM_RESPIRATORY_COLUMNAR_CELLS | 1.81e-04 | 31.85 | 5.95 | 2.52e-03 | 1.21e-01 | 3KRT18, AQP3, KRT8 |
28 |
| FAN_EMBRYONIC_CTX_BRAIN_MYELOID | 1.10e-06 | 15.41 | 5.74 | 3.06e-05 | 7.36e-04 | 7ANXA1, S100A10, CRIP1, S100A4, IFITM2, S100A11, S100A6 |
139 |
| BUSSLINGER_ESOPHAGEAL_LATE_SUPRABASAL_CELLS | 1.15e-06 | 15.29 | 5.70 | 3.06e-05 | 7.72e-04 | 7ANXA1, S100A10, SLPI, AQP3, MAL2, S100A11, LGALS3 |
140 |
| TRAVAGLINI_LUNG_PROXIMAL_CILIATED_CELL | 1.86e-14 | 9.94 | 5.46 | 6.22e-12 | 1.24e-11 | 29ANXA1, CRIP1, CCDC17, ODF3B, PSCA, TMEM190, SMIM22, TPPP3, FOXJ1, DNAAF1, ALDH1L1, RSPH1, TEKT4, PLEKHS1, MORN2, S100A11, CIB1, UCP2, TUBB4B, ITGB4, TTC29, LGALS3, CETN2, SPEF1, EFCAB1, SLC9A3R1, BAIAP3, CYBA, DNAH12 |
1770 |
| TRAVAGLINI_LUNG_CILIATED_CELL | 6.57e-13 | 9.96 | 5.43 | 1.47e-10 | 4.41e-10 | 23ANXA1, CCDC17, ODF3B, TMEM190, SMIM22, TPPP3, FOXJ1, DNAAF1, RSPH1, TEKT4, PLEKHS1, MORN2, B3GNT7, S100A11, CIB1, UCP2, TUBB4B, TTC29, CETN2, SPEF1, EFCAB1, BAIAP3, DNAH12 |
1094 |
| MENON_FETAL_KIDNEY_10_IMMUNE_CELLS | 1.82e-05 | 17.79 | 5.37 | 3.60e-04 | 1.22e-02 | 5S100A4, S100A11, S100A6, B2M, CYBA |
83 |
| DESCARTES_FETAL_LUNG_CILIATED_EPITHELIAL_CELLS | 5.88e-10 | 10.49 | 5.29 | 4.38e-08 | 3.94e-07 | 15CCDC17, ODF3B, TMEM190, TPPP3, FOXJ1, DNAAF1, RSPH1, TEKT4, PLEKHS1, MORN2, TTC29, CETN2, SPEF1, EFCAB1, DNAH12 |
526 |
| CUI_DEVELOPING_HEART_C6_EPICARDIAL_CELL | 8.06e-07 | 12.73 | 5.09 | 2.58e-05 | 5.41e-04 | 8S100A10, KRT18, CRIP1, SLPI, MAL2, S100A6, KRT8, SLC9A3R1 |
195 |
Dowload full table
mSigDB Hallmark Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| HALLMARK_CHOLESTEROL_HOMEOSTASIS | 2.04e-04 | 15.52 | 3.95 | 1.02e-02 | 1.02e-02 | 4MAL2, S100A11, TNFRSF12A, LGALS3 |
74 |
| HALLMARK_APOPTOSIS | 3.63e-03 | 6.92 | 1.79 | 9.08e-02 | 1.82e-01 | 4ANXA1, KRT18, TNFRSF12A, LGALS3 |
161 |
| HALLMARK_INTERFERON_ALPHA_RESPONSE | 6.61e-03 | 8.48 | 1.66 | 9.64e-02 | 3.30e-01 | 3IFITM2, B2M, IFI27 |
97 |
| HALLMARK_ESTROGEN_RESPONSE_EARLY | 7.71e-03 | 5.55 | 1.44 | 9.64e-02 | 3.85e-01 | 4KRT18, AQP3, KRT8, SLC9A3R1 |
200 |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | 4.35e-02 | 4.05 | 0.80 | 3.11e-01 | 1.00e+00 | 3IFITM2, B2M, IFI27 |
200 |
| HALLMARK_P53_PATHWAY | 4.35e-02 | 4.05 | 0.80 | 3.11e-01 | 1.00e+00 | 3S100A10, S100A4, ITGB4 |
200 |
| HALLMARK_HEME_METABOLISM | 4.35e-02 | 4.05 | 0.80 | 3.11e-01 | 1.00e+00 | 3AQP3, ALDH1L1, UCP2 |
200 |
| HALLMARK_ANDROGEN_RESPONSE | 5.93e-02 | 5.31 | 0.62 | 3.71e-01 | 1.00e+00 | 2KRT8, B2M |
100 |
| HALLMARK_COAGULATION | 1.03e-01 | 3.83 | 0.45 | 5.71e-01 | 1.00e+00 | 2ANXA1, S100A1 |
138 |
| HALLMARK_ANGIOGENESIS | 1.34e-01 | 7.29 | 0.18 | 6.68e-01 | 1.00e+00 | 1S100A4 |
36 |
| HALLMARK_IL6_JAK_STAT3_SIGNALING | 2.93e-01 | 2.97 | 0.07 | 1.00e+00 | 1.00e+00 | 1TNFRSF12A |
87 |
| HALLMARK_PROTEIN_SECRETION | 3.17e-01 | 2.68 | 0.07 | 1.00e+00 | 1.00e+00 | 1KRT18 |
96 |
| HALLMARK_DNA_REPAIR | 4.49e-01 | 1.71 | 0.04 | 1.00e+00 | 1.00e+00 | 1CETN2 |
150 |
| HALLMARK_FATTY_ACID_METABOLISM | 4.66e-01 | 1.62 | 0.04 | 1.00e+00 | 1.00e+00 | 1S100A10 |
158 |
| HALLMARK_UV_RESPONSE_UP | 4.66e-01 | 1.62 | 0.04 | 1.00e+00 | 1.00e+00 | 1AQP3 |
158 |
| HALLMARK_IL2_STAT5_SIGNALING | 5.45e-01 | 1.29 | 0.03 | 1.00e+00 | 1.00e+00 | 1S100A1 |
199 |
| HALLMARK_HYPOXIA | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1S100A4 |
200 |
| HALLMARK_ADIPOGENESIS | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1UCP2 |
200 |
| HALLMARK_ESTROGEN_RESPONSE_LATE | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1SLC9A3R1 |
200 |
| HALLMARK_MYOGENESIS | 5.47e-01 | 1.28 | 0.03 | 1.00e+00 | 1.00e+00 | 1ITGB4 |
200 |
Dowload full table
KEGG Pathways:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| KEGG_PATHOGENIC_ESCHERICHIA_COLI_INFECTION | 2.07e-02 | 9.64 | 1.11 | 1.00e+00 | 1.00e+00 | 2KRT18, TUBB4B |
56 |
| KEGG_GLYCOSAMINOGLYCAN_BIOSYNTHESIS_KERATAN_SULFATE | 5.81e-02 | 18.20 | 0.42 | 1.00e+00 | 1.00e+00 | 1B3GNT7 |
15 |
| KEGG_ONE_CARBON_POOL_BY_FOLATE | 6.56e-02 | 15.93 | 0.37 | 1.00e+00 | 1.00e+00 | 1ALDH1L1 |
17 |
| KEGG_NUCLEOTIDE_EXCISION_REPAIR | 1.61e-01 | 5.93 | 0.14 | 1.00e+00 | 1.00e+00 | 1CETN2 |
44 |
| KEGG_VASOPRESSIN_REGULATED_WATER_REABSORPTION | 1.61e-01 | 5.93 | 0.14 | 1.00e+00 | 1.00e+00 | 1AQP3 |
44 |
| KEGG_LEISHMANIA_INFECTION | 2.49e-01 | 3.59 | 0.09 | 1.00e+00 | 1.00e+00 | 1CYBA |
72 |
| KEGG_ARRHYTHMOGENIC_RIGHT_VENTRICULAR_CARDIOMYOPATHY_ARVC | 2.55e-01 | 3.49 | 0.09 | 1.00e+00 | 1.00e+00 | 1ITGB4 |
74 |
| KEGG_HYPERTROPHIC_CARDIOMYOPATHY_HCM | 2.81e-01 | 3.11 | 0.08 | 1.00e+00 | 1.00e+00 | 1ITGB4 |
83 |
| KEGG_ECM_RECEPTOR_INTERACTION | 2.84e-01 | 3.07 | 0.08 | 1.00e+00 | 1.00e+00 | 1ITGB4 |
84 |
| KEGG_ANTIGEN_PROCESSING_AND_PRESENTATION | 2.95e-01 | 2.93 | 0.07 | 1.00e+00 | 1.00e+00 | 1B2M |
88 |
| KEGG_DILATED_CARDIOMYOPATHY | 3.01e-01 | 2.87 | 0.07 | 1.00e+00 | 1.00e+00 | 1ITGB4 |
90 |
| KEGG_GAP_JUNCTION | 3.01e-01 | 2.87 | 0.07 | 1.00e+00 | 1.00e+00 | 1TUBB4B |
90 |
| KEGG_LEUKOCYTE_TRANSENDOTHELIAL_MIGRATION | 3.69e-01 | 2.22 | 0.05 | 1.00e+00 | 1.00e+00 | 1CYBA |
116 |
| KEGG_FOCAL_ADHESION | 5.45e-01 | 1.29 | 0.03 | 1.00e+00 | 1.00e+00 | 1ITGB4 |
199 |
| KEGG_REGULATION_OF_ACTIN_CYTOSKELETON | 5.70e-01 | 1.20 | 0.03 | 1.00e+00 | 1.00e+00 | 1ITGB4 |
213 |
| KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION | 1.00e+00 | 0.97 | 0.02 | 1.00e+00 | 1.00e+00 | 1TNFRSF12A |
265 |
| KEGG_N_GLYCAN_BIOSYNTHESIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
46 |
| KEGG_OTHER_GLYCAN_DEGRADATION | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
16 |
| KEGG_O_GLYCAN_BIOSYNTHESIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
30 |
| KEGG_GLYCOSAMINOGLYCAN_DEGRADATION | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
21 |
Dowload full table
CHR Positional Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| chr1q21 | 1.72e-02 | 3.59 | 1.11 | 1.00e+00 | 1.00e+00 | 5S100A10, S100A4, S100A1, S100A11, S100A6 |
392 |
| chr17q25 | 1.09e-01 | 2.71 | 0.54 | 1.00e+00 | 1.00e+00 | 3FOXJ1, ITGB4, SLC9A3R1 |
297 |
| chr16q24 | 9.30e-02 | 4.07 | 0.47 | 1.00e+00 | 1.00e+00 | 2DNAAF1, CYBA |
130 |
| chr16p13 | 2.07e-01 | 1.98 | 0.39 | 1.00e+00 | 1.00e+00 | 3SMIM22, TNFRSF12A, BAIAP3 |
407 |
| chr15q21 | 1.40e-01 | 3.16 | 0.37 | 1.00e+00 | 1.00e+00 | 2DUOX2, B2M |
167 |
| chr9q34 | 3.42e-01 | 1.69 | 0.20 | 1.00e+00 | 1.00e+00 | 2TUBB4B, GFI1B |
311 |
| chr8q24 | 3.56e-01 | 1.63 | 0.19 | 1.00e+00 | 1.00e+00 | 2PSCA, MAL2 |
321 |
| chr12q13 | 6.71e-01 | 1.29 | 0.15 | 1.00e+00 | 1.00e+00 | 2KRT18, KRT8 |
407 |
| chr14q32 | 1.00e+00 | 0.96 | 0.11 | 1.00e+00 | 1.00e+00 | 2CRIP1, IFI27 |
546 |
| chr8q11 | 2.43e-01 | 3.70 | 0.09 | 1.00e+00 | 1.00e+00 | 1EFCAB1 |
70 |
| chr14q22 | 3.01e-01 | 2.87 | 0.07 | 1.00e+00 | 1.00e+00 | 1LGALS3 |
90 |
| chr2p22 | 3.23e-01 | 2.63 | 0.06 | 1.00e+00 | 1.00e+00 | 1MORN2 |
98 |
| chr20p13 | 3.72e-01 | 2.20 | 0.05 | 1.00e+00 | 1.00e+00 | 1SPEF1 |
117 |
| chr19q13 | 4.39e-01 | 0.45 | 0.05 | 1.00e+00 | 1.00e+00 | 2TMEM190, MIA |
1165 |
| chr3p14 | 3.84e-01 | 2.11 | 0.05 | 1.00e+00 | 1.00e+00 | 1DNAH12 |
122 |
| chr10q25 | 3.94e-01 | 2.04 | 0.05 | 1.00e+00 | 1.00e+00 | 1PLEKHS1 |
126 |
| chr3q21 | 4.22e-01 | 1.86 | 0.05 | 1.00e+00 | 1.00e+00 | 1ALDH1L1 |
138 |
| chr2q11 | 4.40e-01 | 1.76 | 0.04 | 1.00e+00 | 1.00e+00 | 1TEKT4 |
146 |
| chr4q31 | 4.64e-01 | 1.64 | 0.04 | 1.00e+00 | 1.00e+00 | 1TTC29 |
157 |
| chr9p13 | 4.90e-01 | 1.51 | 0.04 | 1.00e+00 | 1.00e+00 | 1AQP3 |
170 |
Dowload full table
Transcription Factor Targets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GREB1_TARGET_GENES | 7.05e-03 | 2.95 | 1.25 | 1.00e+00 | 1.00e+00 | 9S100A10, S100A4, PSCA, KCTD11, DNAAF1, S100A11, TUBB4B, KRT8, BAIAP3 |
941 |
| BACH1_01 | 1.91e-02 | 4.20 | 1.09 | 1.00e+00 | 1.00e+00 | 4S100A10, TNFRSF12A, KRT8, ITGB4 |
263 |
| CYTAGCAAY_UNKNOWN | 2.15e-02 | 5.39 | 1.06 | 1.00e+00 | 1.00e+00 | 3ANXA1, TMEM190, TPPP3 |
151 |
| ZNF133_TARGET_GENES | 2.26e-02 | 5.28 | 1.04 | 1.00e+00 | 1.00e+00 | 3KRT18, KRT8, IFFO2 |
154 |
| RFX1_02 | 2.49e-02 | 3.85 | 1.00 | 1.00e+00 | 1.00e+00 | 4DNAAF1, MORN2, SPEF1, BAIAP3 |
286 |
| MYBL1_TARGET_GENES | 4.35e-02 | 4.05 | 0.80 | 1.00e+00 | 1.00e+00 | 3KRT18, KRT8, EFCAB1 |
200 |
| FOXJ2_TARGET_GENES | 7.30e-02 | 2.37 | 0.73 | 1.00e+00 | 1.00e+00 | 5TPPP3, FOXJ1, TUBB4B, KRT8, CETN2 |
591 |
| ZBTB7B_TARGET_GENES | 2.07e-01 | 1.62 | 0.72 | 1.00e+00 | 1.00e+00 | 10FOXJ1, DNAAF1, RSPH1, MORN2, UCP2, TUBB4B, TTC29, CETN2, SPEF1, DNAH12 |
1935 |
| RYTTCCTG_ETS2_B | 1.23e-01 | 1.84 | 0.70 | 1.00e+00 | 1.00e+00 | 7SLPI, AQP3, PLEKHS1, B3GNT7, CIB1, MIA, SLC9A3R1 |
1112 |
| ZFP91_TARGET_GENES | 2.37e-01 | 1.66 | 0.67 | 1.00e+00 | 1.00e+00 | 8KCTD11, TEKT4, GFI1B, TNFRSF12A, KRT8, B2M, IFFO2, ZBTB22 |
1442 |
| TRIP13_TARGET_GENES | 5.52e-02 | 5.54 | 0.64 | 1.00e+00 | 1.00e+00 | 2PSCA, GFI1B |
96 |
| NAB2_TARGET_GENES | 2.47e-01 | 1.58 | 0.64 | 1.00e+00 | 1.00e+00 | 8S100A10, RGCC, ODF3B, DNAAF1, CIB1, TNFRSF12A, KRT8, BAIAP3 |
1512 |
| HNF4_Q6 | 8.87e-02 | 2.98 | 0.59 | 1.00e+00 | 1.00e+00 | 3SLPI, AQP3, SLC9A3R1 |
271 |
| ACTWSNACTNY_UNKNOWN | 6.57e-02 | 5.01 | 0.58 | 1.00e+00 | 1.00e+00 | 2SPEF1, SLC9A3R1 |
106 |
| ATXN7L3_TARGET_GENES | 9.62e-02 | 2.87 | 0.57 | 1.00e+00 | 1.00e+00 | 3S100A10, KRT18, S100A6 |
281 |
| TATA_C | 9.93e-02 | 2.83 | 0.56 | 1.00e+00 | 1.00e+00 | 3ANXA1, S100A10, DUOX2 |
285 |
| NKX2_3_TARGET_GENES | 1.50e-01 | 2.03 | 0.53 | 1.00e+00 | 1.00e+00 | 4ODF3B, S100A11, TUBB4B, KRT8 |
539 |
| PSMB5_TARGET_GENES | 1.17e-01 | 2.62 | 0.52 | 1.00e+00 | 1.00e+00 | 3S100A1, S100A6, TUBB4B |
307 |
| ZNF184_TARGET_GENES | 4.86e-01 | 1.37 | 0.52 | 1.00e+00 | 1.00e+00 | 7CIB1, UCP2, GFI1B, KRT8, CETN2, BAIAP3, ZBTB22 |
1496 |
| FOXN3_TARGET_GENES | 4.87e-01 | 1.36 | 0.52 | 1.00e+00 | 1.00e+00 | 7FOXJ1, DNAAF1, RSPH1, MORN2, B3GNT7, ITGB4, SPEF1 |
1502 |
Dowload full table
GO Biological Processes:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GOBP_HEPATOCYTE_APOPTOTIC_PROCESS | 1.37e-03 | 43.30 | 4.59 | 1.00e+00 | 1.00e+00 | 2KRT18, KRT8 |
14 |
| GOBP_MICROTUBULE_BUNDLE_FORMATION | 6.16e-05 | 13.61 | 4.13 | 1.19e-01 | 4.61e-01 | 5TPPP3, FOXJ1, DNAAF1, RSPH1, SPEF1 |
107 |
| GOBP_HEPATOCYTE_DIFFERENTIATION | 1.80e-03 | 37.11 | 3.99 | 1.00e+00 | 1.00e+00 | 2ANXA1, KRT18 |
16 |
| GOBP_AXONEME_ASSEMBLY | 2.26e-04 | 15.09 | 3.84 | 2.82e-01 | 1.00e+00 | 4FOXJ1, DNAAF1, RSPH1, SPEF1 |
76 |
| GOBP_HYDROGEN_PEROXIDE_BIOSYNTHETIC_PROCESS | 2.03e-03 | 34.68 | 3.75 | 1.00e+00 | 1.00e+00 | 2DUOX2, CYBA |
17 |
| GOBP_RENAL_ABSORPTION | 2.28e-03 | 32.49 | 3.53 | 1.00e+00 | 1.00e+00 | 2AQP3, SLC9A3R1 |
18 |
| GOBP_LEFT_RIGHT_PATTERN_FORMATION | 2.54e-03 | 30.58 | 3.34 | 1.00e+00 | 1.00e+00 | 2FOXJ1, DNAAF1 |
19 |
| GOBP_CILIUM_ORGANIZATION | 2.22e-05 | 6.88 | 2.92 | 1.19e-01 | 1.66e-01 | 9FOXJ1, DNAAF1, RSPH1, TEKT4, TUBB4B, TTC29, CETN2, SPEF1, SLC9A3R1 |
408 |
| GOBP_EXTRINSIC_APOPTOTIC_SIGNALING_PATHWAY | 1.92e-04 | 8.08 | 2.78 | 2.82e-01 | 1.00e+00 | 6KRT18, CIB1, TNFRSF12A, KRT8, IFI27, LGALS3 |
217 |
| GOBP_MOTILE_CILIUM_ASSEMBLY | 3.72e-03 | 24.78 | 2.74 | 1.00e+00 | 1.00e+00 | 2FOXJ1, DNAAF1 |
23 |
| GOBP_ESTABLISHMENT_OR_MAINTENANCE_OF_MONOPOLAR_CELL_POLARITY | 3.72e-03 | 24.78 | 2.74 | 1.00e+00 | 1.00e+00 | 2FOXJ1, SLC9A3R1 |
23 |
| GOBP_CILIUM_MOVEMENT | 4.46e-04 | 8.73 | 2.67 | 4.17e-01 | 1.00e+00 | 5DNAAF1, TEKT4, TTC29, SPEF1, SLC9A3R1 |
164 |
| GOBP_MICROTUBULE_BASED_PROCESS | 4.80e-05 | 4.74 | 2.25 | 1.19e-01 | 3.59e-01 | 12TPPP3, FOXJ1, DNAAF1, RSPH1, TEKT4, CIB1, TUBB4B, TTC29, CETN2, SPEF1, SLC9A3R1, DNAH12 |
845 |
| GOBP_REGULATION_OF_MICROTUBULE_DEPOLYMERIZATION | 5.87e-03 | 19.27 | 2.16 | 1.00e+00 | 1.00e+00 | 2CIB1, SPEF1 |
29 |
| GOBP_HEPATICOBILIARY_SYSTEM_DEVELOPMENT | 2.21e-03 | 7.99 | 2.06 | 1.00e+00 | 1.00e+00 | 4ANXA1, KRT18, DNAAF1, UCP2 |
140 |
| GOBP_CELL_PROJECTION_ASSEMBLY | 3.56e-04 | 4.68 | 1.99 | 3.81e-01 | 1.00e+00 | 9FOXJ1, DNAAF1, RSPH1, TEKT4, TUBB4B, ITGB4, CETN2, SPEF1, SLC9A3R1 |
595 |
| GOBP_CYTOSKELETON_ORGANIZATION | 6.39e-05 | 3.93 | 1.99 | 1.19e-01 | 4.78e-01 | 15ANXA1, S100A10, KRT18, RGCC, SMIM22, TPPP3, FOXJ1, DNAAF1, RSPH1, CIB1, TUBB4B, KRT8, CETN2, SPEF1, SLC9A3R1 |
1380 |
| GOBP_POSITIVE_REGULATION_OF_CELL_MIGRATION_INVOLVED_IN_SPROUTING_ANGIOGENESIS | 8.00e-03 | 16.26 | 1.84 | 1.00e+00 | 1.00e+00 | 2ANXA1, CIB1 |
34 |
| GOBP_EXTRACELLULAR_TRANSPORT | 8.47e-03 | 15.77 | 1.78 | 1.00e+00 | 1.00e+00 | 2DNAAF1, SLC9A3R1 |
35 |
| GOBP_SUPEROXIDE_ANION_GENERATION | 8.47e-03 | 15.77 | 1.78 | 1.00e+00 | 1.00e+00 | 2DUOX2, CYBA |
35 |
Dowload full table
Immunological Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GSE11057_NAIVE_VS_EFF_MEMORY_CD4_TCELL_DN | 8.70e-07 | 12.60 | 5.04 | 2.20e-03 | 4.24e-03 | 8ANXA1, CRIP1, S100A4, AQP3, S100A11, IFI27, LGALS3, SLC9A3R1 |
197 |
| GSE11057_NAIVE_VS_MEMORY_CD4_TCELL_DN | 9.03e-07 | 12.53 | 5.01 | 2.20e-03 | 4.40e-03 | 8ANXA1, CRIP1, S100A4, AQP3, S100A11, TUBB4B, LGALS3, SLC9A3R1 |
198 |
| KAECH_NAIVE_VS_DAY8_EFF_CD8_TCELL_DN | 1.19e-05 | 10.54 | 3.95 | 1.45e-02 | 5.79e-02 | 7ANXA1, S100A10, CRIP1, S100A4, S100A11, S100A6, LGALS3 |
200 |
| GSE23568_CTRL_TRANSDUCED_VS_WT_CD8_TCELL_UP | 1.19e-05 | 10.54 | 3.95 | 1.45e-02 | 5.79e-02 | 7ANXA1, S100A10, CRIP1, S100A4, S100A11, S100A6, LGALS3 |
200 |
| GSE13522_WT_VS_IFNAR_KO_SKIN_DN | 6.36e-05 | 9.97 | 3.42 | 2.74e-02 | 3.10e-01 | 6S100A10, CRIP1, S100A4, S100A11, S100A6, LGALS3 |
177 |
| GSE15330_LYMPHOID_MULTIPOTENT_VS_MEGAKARYOCYTE_ERYTHROID_PROGENITOR_IKAROS_KO_DN | 6.56e-05 | 9.91 | 3.41 | 2.74e-02 | 3.20e-01 | 6ANXA1, S100A10, CRIP1, S100A4, S100A11, S100A6 |
178 |
| GSE40274_CTRL_VS_FOXP3_AND_GATA1_TRANSDUCED_ACTIVATED_CD4_TCELL_UP | 2.24e-04 | 10.21 | 3.11 | 4.74e-02 | 1.00e+00 | 5S100A10, KCTD11, SLPI, S100A11, S100A6 |
141 |
| GSE22886_NAIVE_VS_IGG_IGA_MEMORY_BCELL_DN | 1.11e-04 | 8.97 | 3.09 | 2.74e-02 | 5.40e-01 | 6S100A10, CRIP1, S100A4, S100A6, TUBB4B, BAIAP3 |
196 |
| GSE11057_NAIVE_VS_CENT_MEMORY_CD4_TCELL_DN | 1.20e-04 | 8.83 | 3.04 | 2.74e-02 | 5.86e-01 | 6ANXA1, CRIP1, S100A4, AQP3, LGALS3, IFFO2 |
199 |
| GSE29618_MONOCYTE_VS_PDC_UP | 1.20e-04 | 8.83 | 3.04 | 2.74e-02 | 5.86e-01 | 6ANXA1, S100A10, S100A4, IFITM2, S100A11, LGALS3 |
199 |
| GSE9650_NAIVE_VS_EFF_CD8_TCELL_DN | 1.20e-04 | 8.83 | 3.04 | 2.74e-02 | 5.86e-01 | 6ANXA1, S100A10, S100A4, S100A11, S100A6, LGALS3 |
199 |
| KAECH_NAIVE_VS_DAY15_EFF_CD8_TCELL_DN | 1.24e-04 | 8.78 | 3.02 | 2.74e-02 | 6.03e-01 | 6ANXA1, S100A10, S100A4, S100A11, S100A6, LGALS3 |
200 |
| GSE15930_STIM_VS_STIM_AND_TRICHOSTATINA_48H_CD8_T_CELL_UP | 1.24e-04 | 8.78 | 3.02 | 2.74e-02 | 6.03e-01 | 6S100A10, S100A4, S100A1, S100A11, S100A6, LGALS3 |
200 |
| GSE29618_PDC_VS_MDC_DAY7_FLU_VACCINE_DN | 1.24e-04 | 8.78 | 3.02 | 2.74e-02 | 6.03e-01 | 6ANXA1, S100A10, RGCC, S100A4, IFITM2, LGALS3 |
200 |
| GSE34205_HEALTHY_VS_FLU_INF_INFANT_PBMC_DN | 1.24e-04 | 8.78 | 3.02 | 2.74e-02 | 6.03e-01 | 6IFITM2, ODF3B, S100A11, S100A6, B2M, IFI27 |
200 |
| GSE7460_FOXP3_MUT_VS_HET_ACT_TCONV_UP | 1.24e-04 | 8.78 | 3.02 | 2.74e-02 | 6.03e-01 | 6KRT18, IFITM2, RSPH1, S100A6, UCP2, LGALS3 |
200 |
| GSE1925_CTRL_VS_IFNG_PRIMED_MACROPHAGE_24H_IFNG_STIM_DN | 1.24e-04 | 8.78 | 3.02 | 2.74e-02 | 6.03e-01 | 6TPPP3, S100A11, S100A6, B2M, IFI27, SLC9A3R1 |
200 |
| GSE18203_CTRL_VS_INTRATUMORAL_CPG_INJ_MC38_TUMOR_DN | 1.24e-04 | 8.78 | 3.02 | 2.74e-02 | 6.03e-01 | 6S100A10, S100A4, S100A1, S100A11, S100A6, LGALS3 |
200 |
| GSE18893_TCONV_VS_TREG_24H_CULTURE_UP | 1.24e-04 | 8.78 | 3.02 | 2.74e-02 | 6.03e-01 | 6ANXA1, S100A10, CRIP1, S100A4, S100A6, LGALS3 |
200 |
| GSE23568_ID3_TRANSDUCED_VS_ID3_KO_CD8_TCELL_UP | 1.24e-04 | 8.78 | 3.02 | 2.74e-02 | 6.03e-01 | 6S100A10, RGCC, S100A4, S100A6, B2M, IFI27 |
200 |
Top Ranked Transcription Factors for this Gene Expression Program:
| Gene Symbol | TF Rank | DNA Binding Domain | Motif Status | IUPAC PWM | GTEx | DepMap | Decartes |
|---|---|---|---|---|---|---|---|
| RGCC | 5 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| CCDC17 | 9 | Yes | Likely to be sequence specific TF | Monomer or homomultimer | No motif | Single C2H2 domain | None |
| FOXJ1 | 18 | Yes | Known motif | Monomer or homomultimer | 100 perc ID - in vitro | None | None |
| CIB1 | 30 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| GFI1B | 33 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| ZBTB22 | 50 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| CEBPB | 55 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| MACC1 | 62 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| KLF5 | 64 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | Binds as a monomer and as a dimer (PMID: 25575120). |
| TNFAIP3 | 67 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Inhibits NFKB-activation. No evidence for direct binding to DNA |
| THAP11 | 102 | Yes | Likely to be sequence specific TF | Monomer or homomultimer | No motif | None | (PMID: 26876175) has a ChIP-seq motif with GGGARWTGTAGT consensus. THAP11 is an atypical zinc-finger paralogous to THAP1, which has a crystal structure (PDB: 2KO0) where the protein contacts bases TGGGCA |
| IRF9 | 129 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| HSBP1 | 132 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Protein that has been shown to bind to HSF1 leading to inhibition of its trimerization and DNA-binding( PMID: 9649501). |
| HEY1 | 140 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| SGK1 | 165 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| PLAGL2 | 196 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| NFKBIA | 197 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | This is I kappa B alpha, which interacts with NFkappaB. No DBD, and evidence indicates that it is not a TF. |
| TAF10 | 209 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | General transcription factor |
| ERF | 210 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| ZMAT2 | 229 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Possibly an RBP based on presence of a U1 ZF domain |
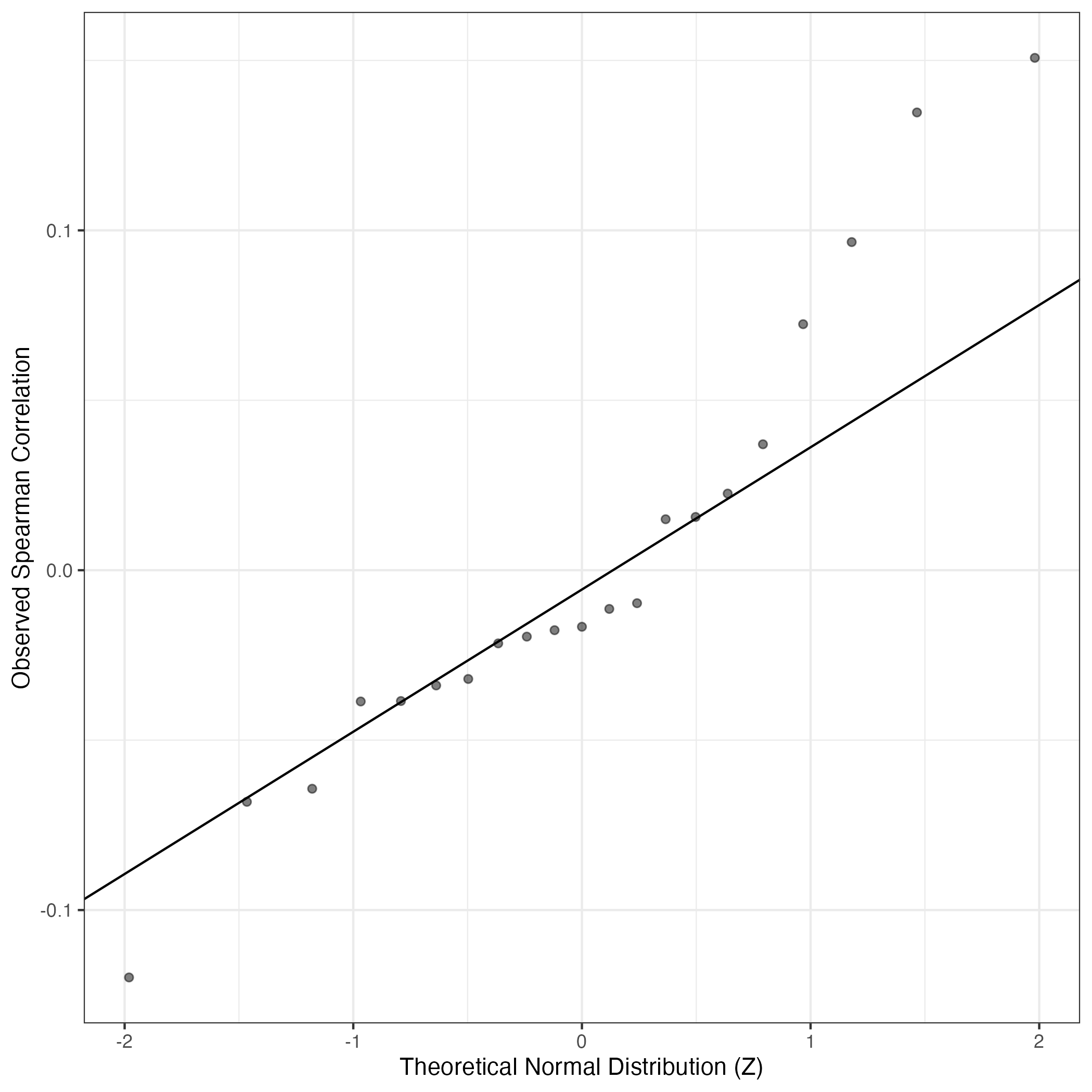
QQ Plot showing correlations with other Gene Expression Programs in this dataset, calculated by Spearman correlation:
Interactive QQ-plot of gene loadings:
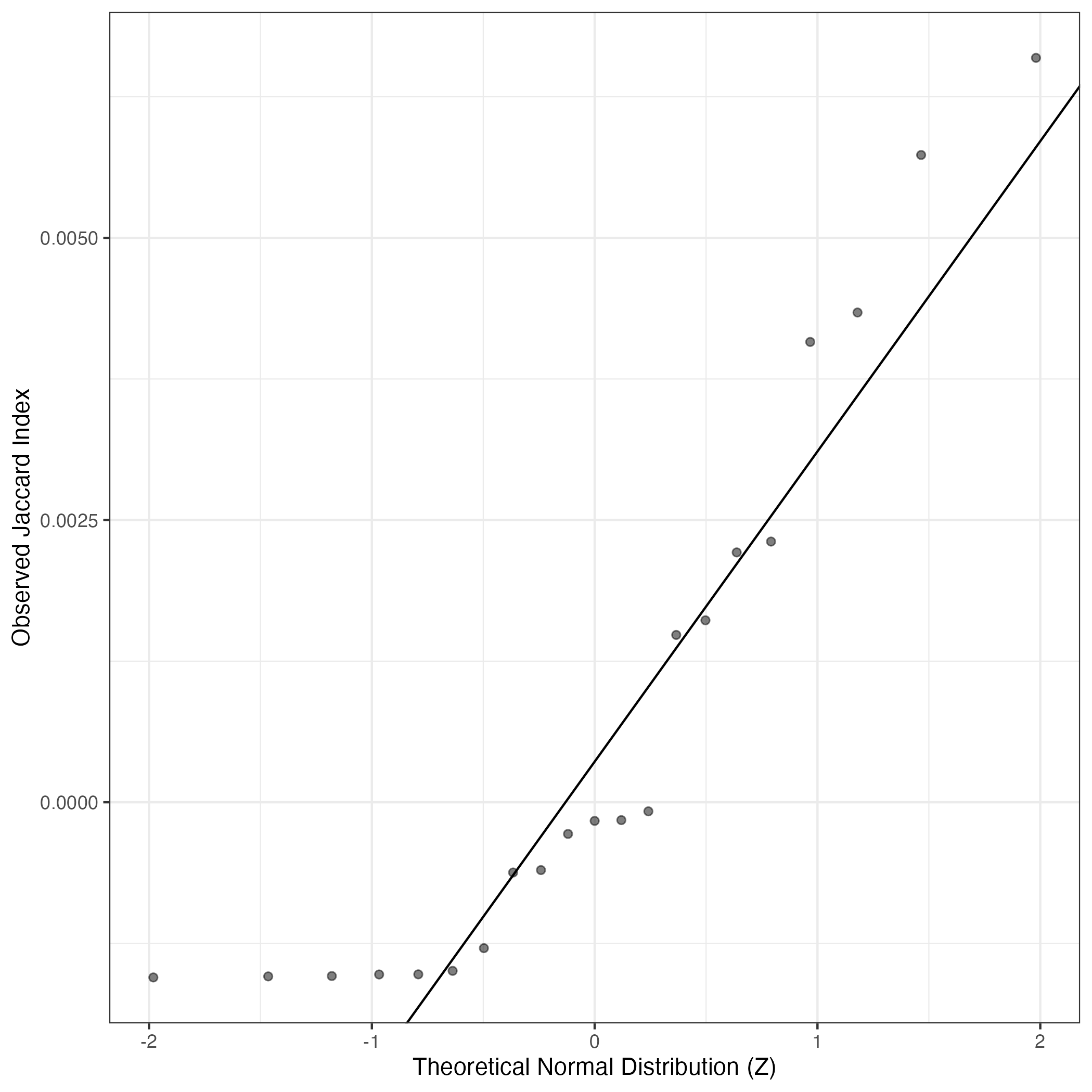
A similar QQ-plot as above, but only for instances where the H value is e.g. > 25, i.e. we are confident that the expression program is active above noise. Agreemenet between these binary vectors is tested using the Jaccard Index, with the P-values calculated by an exact test:
Interactive QQ-plot:
Singler cell type annotations for the top 50 cells on this program.
## Error in 1:nrow(topCellsAnnotMat): argument of length 0
| Cell ID | Singler label | Singler Delta | Activity Score | Top Singler Raw Scores |
|---|---|---|---|---|
| SJNBL046_sn_TCAGATGGTGCACCAC-1 | Myelocyte | 0.04 | 614.5 | Raw Scores |
Below shows the significant enrichments of this GEP for literature curated gene lists
This data was procured from existing single cell RNA-seq maps of neuroblastoma or related relevant data.
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by the P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Antigen Presenting CAF
These marker genes were curated across cancer subtypes in multiple organ systems as reviewed in Lavie et. al. (PMID 35883004) and contain antigen presenting specific CAF genes:
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.21e-05
Mean rank of genes in gene set: 1862.33
Rank on gene expression program of genes in gene set:
## Error in data.frame(Genes = geneCardsLinkSmallGsets, Weight = theseGenes, : object 'countHighScoringCells' not found