Program: 38. M1 Macrophage.
Submit a comment on this gene expression program’s interpretation: CLICK
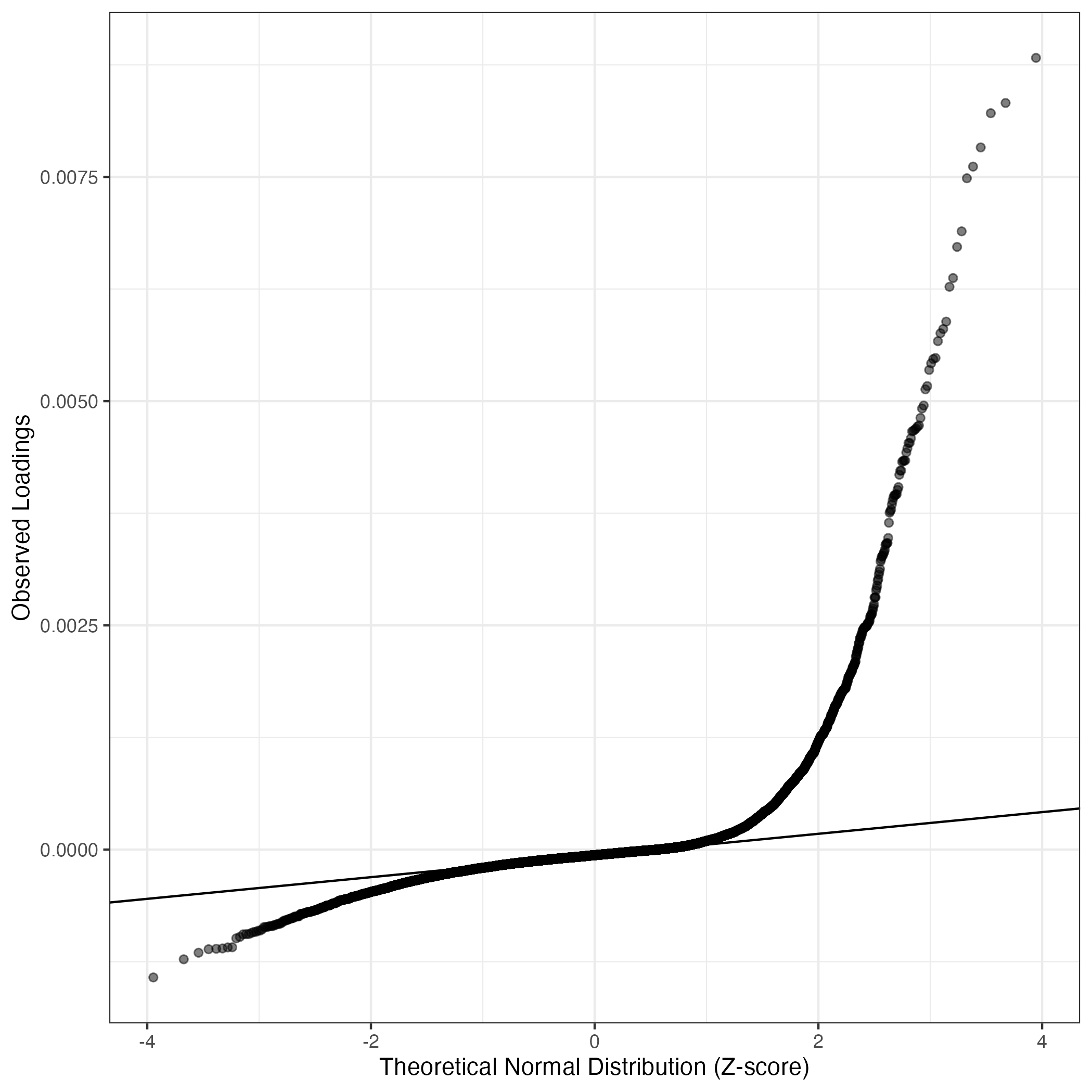
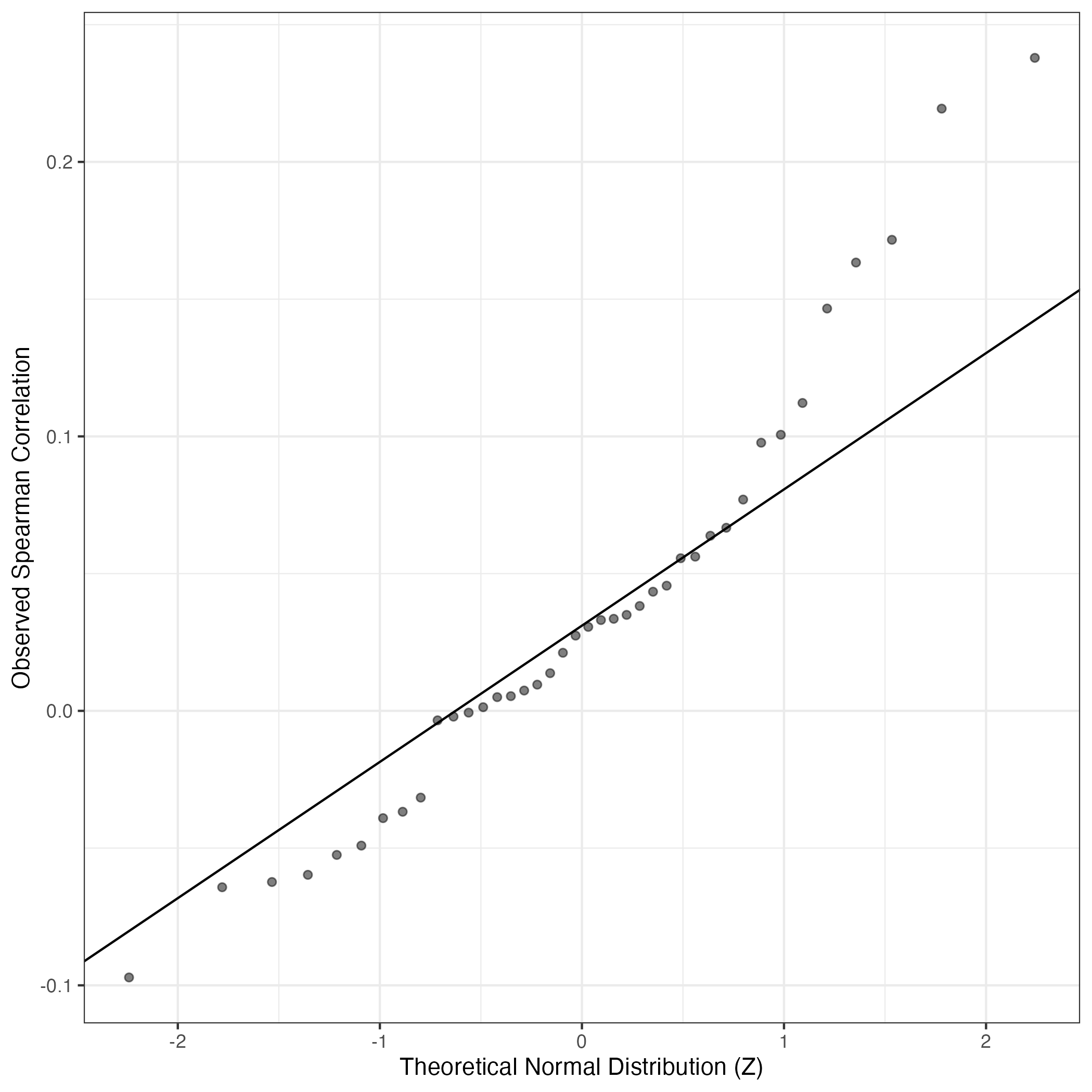
QQ-plot of gene loadings, averaged over both independent splits of the data
This plot highlights the relative contribution of each gene to the GEP
Top genes driving this program.
Note: Decartes website is buggy, try refreshing. Also, Decartes fetal adrenal data have been collected at specific time points (89-122 days), all possible cell types of interest may not be represented, do not overinterpret.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
| Gene | Loading | Gene.Name | GTEx | DepMap | Descartes | Mean.Counts | Mean.Tpm | |
|---|---|---|---|---|---|---|---|---|
| 1 | IER2 | 0.0088277 | immediate early response 2 | GTEx | DepMap | Descartes | 13.06 | 1000.29 |
| 2 | ATF3 | 0.0083247 | activating transcription factor 3 | GTEx | DepMap | Descartes | 6.63 | 688.64 |
| 3 | IER3 | 0.0082105 | immediate early response 3 | GTEx | DepMap | Descartes | 17.77 | 2597.22 |
| 4 | NFKBIA | 0.0078290 | NFKB inhibitor alpha | GTEx | DepMap | Descartes | 27.97 | 3823.18 |
| 5 | GADD45B | 0.0076151 | growth arrest and DNA damage inducible beta | GTEx | DepMap | Descartes | 10.87 | 1247.25 |
| 6 | EIF4A3 | 0.0074852 | eukaryotic translation initiation factor 4A3 | GTEx | DepMap | Descartes | 3.64 | 211.04 |
| 7 | FOSB | 0.0068932 | FosB proto-oncogene, AP-1 transcription factor subunit | GTEx | DepMap | Descartes | 9.58 | 594.75 |
| 8 | DUSP1 | 0.0067197 | dual specificity phosphatase 1 | GTEx | DepMap | Descartes | 19.94 | 2221.59 |
| 9 | NR4A2 | 0.0063727 | nuclear receptor subfamily 4 group A member 2 | GTEx | DepMap | Descartes | 5.88 | 391.79 |
| 10 | CD83 | 0.0062750 | CD83 molecule | GTEx | DepMap | Descartes | 6.97 | 694.11 |
| 11 | FOS | 0.0058866 | Fos proto-oncogene, AP-1 transcription factor subunit | GTEx | DepMap | Descartes | 31.24 | 3242.58 |
| 12 | PPP1R15A | 0.0058043 | protein phosphatase 1 regulatory subunit 15A | GTEx | DepMap | Descartes | 6.44 | 608.10 |
| 13 | CD5L | 0.0057570 | CD5 molecule like | GTEx | DepMap | Descartes | 1.32 | 105.92 |
| 14 | SQSTM1 | 0.0056690 | sequestosome 1 | GTEx | DepMap | Descartes | 3.86 | 290.72 |
| 15 | IFI30 | 0.0054815 | IFI30 lysosomal thiol reductase | GTEx | DepMap | Descartes | 1.21 | 115.79 |
| 16 | STX11 | 0.0054694 | syntaxin 11 | GTEx | DepMap | Descartes | 1.25 | 46.20 |
| 17 | JUNB | 0.0054216 | JunB proto-oncogene, AP-1 transcription factor subunit | GTEx | DepMap | Descartes | 26.29 | 3101.13 |
| 18 | JDP2 | 0.0053479 | Jun dimerization protein 2 | GTEx | DepMap | Descartes | 0.48 | 18.41 |
| 19 | FGL2 | 0.0051679 | fibrinogen like 2 | GTEx | DepMap | Descartes | 3.13 | 134.57 |
| 20 | VCAM1 | 0.0051315 | vascular cell adhesion molecule 1 | GTEx | DepMap | Descartes | 0.61 | 28.99 |
| 21 | ICAM1 | 0.0049514 | intercellular adhesion molecule 1 | GTEx | DepMap | Descartes | 2.34 | 168.80 |
| 22 | SAT1 | 0.0049181 | spermidine/spermine N1-acetyltransferase 1 | GTEx | DepMap | Descartes | 37.79 | 6868.21 |
| 23 | DUSP2 | 0.0048128 | dual specificity phosphatase 2 | GTEx | DepMap | Descartes | 9.81 | 1294.82 |
| 24 | KLF4 | 0.0047289 | Kruppel like factor 4 | GTEx | DepMap | Descartes | 2.16 | 157.54 |
| 25 | HERPUD1 | 0.0047169 | homocysteine inducible ER protein with ubiquitin like domain 1 | GTEx | DepMap | Descartes | 4.68 | 354.36 |
| 26 | NEAT1 | 0.0046948 | nuclear paraspeckle assembly transcript 1 | GTEx | DepMap | Descartes | 13.43 | 129.48 |
| 27 | KLF6 | 0.0046827 | Kruppel like factor 6 | GTEx | DepMap | Descartes | 9.10 | 471.40 |
| 28 | PLA2G7 | 0.0046684 | phospholipase A2 group VII | GTEx | DepMap | Descartes | 2.11 | 184.96 |
| 29 | ZFAND5 | 0.0046636 | zinc finger AN1-type containing 5 | GTEx | DepMap | Descartes | 2.99 | 103.32 |
| 30 | MS4A4A | 0.0045853 | membrane spanning 4-domains A4A | GTEx | DepMap | Descartes | 2.94 | 310.97 |
| 31 | MMP19 | 0.0045348 | matrix metallopeptidase 19 | GTEx | DepMap | Descartes | 0.86 | 50.90 |
| 32 | TNFAIP2 | 0.0045326 | TNF alpha induced protein 2 | GTEx | DepMap | Descartes | 1.34 | 55.38 |
| 33 | ARL4C | 0.0044734 | ADP ribosylation factor like GTPase 4C | GTEx | DepMap | Descartes | 4.21 | 221.49 |
| 34 | JUN | 0.0044268 | Jun proto-oncogene, AP-1 transcription factor subunit | GTEx | DepMap | Descartes | 13.19 | 885.53 |
| 35 | GLA | 0.0043379 | galactosidase alpha | GTEx | DepMap | Descartes | 1.10 | 112.77 |
| 36 | BTG2 | 0.0043367 | BTG anti-proliferation factor 2 | GTEx | DepMap | Descartes | 5.69 | 478.61 |
| 37 | SERTAD1 | 0.0043350 | SERTA domain containing 1 | GTEx | DepMap | Descartes | 1.70 | 198.94 |
| 38 | MAFF | 0.0043234 | MAF bZIP transcription factor F | GTEx | DepMap | Descartes | 1.62 | 108.57 |
| 39 | RILPL2 | 0.0042253 | Rab interacting lysosomal protein like 2 | GTEx | DepMap | Descartes | 2.11 | 68.66 |
| 40 | NR4A1 | 0.0042251 | nuclear receptor subfamily 4 group A member 1 | GTEx | DepMap | Descartes | 3.55 | 164.11 |
| 41 | NAMPT | 0.0041795 | nicotinamide phosphoribosyltransferase | GTEx | DepMap | Descartes | 4.29 | 205.13 |
| 42 | SMPDL3A | 0.0040412 | sphingomyelin phosphodiesterase acid like 3A | GTEx | DepMap | Descartes | 0.85 | 88.88 |
| 43 | TIPARP | 0.0040105 | TCDD inducible poly(ADP-ribose) polymerase | GTEx | DepMap | Descartes | 1.08 | 66.89 |
| 44 | LMNA | 0.0039626 | lamin A/C | GTEx | DepMap | Descartes | 3.74 | 242.94 |
| 45 | SLC1A3 | 0.0039586 | solute carrier family 1 member 3 | GTEx | DepMap | Descartes | 0.50 | 27.11 |
| 46 | MAFB | 0.0039516 | MAF bZIP transcription factor B | GTEx | DepMap | Descartes | 2.47 | 160.95 |
| 47 | PHACTR1 | 0.0039498 | phosphatase and actin regulator 1 | GTEx | DepMap | Descartes | 2.12 | 83.79 |
| 48 | NABP1 | 0.0039254 | nucleic acid binding protein 1 | GTEx | DepMap | Descartes | 0.97 | NA |
| 49 | PPP1R15B | 0.0038901 | protein phosphatase 1 regulatory subunit 15B | GTEx | DepMap | Descartes | 1.08 | 45.00 |
| 50 | C5AR1 | 0.0038479 | complement C5a receptor 1 | GTEx | DepMap | Descartes | 1.96 | 179.49 |
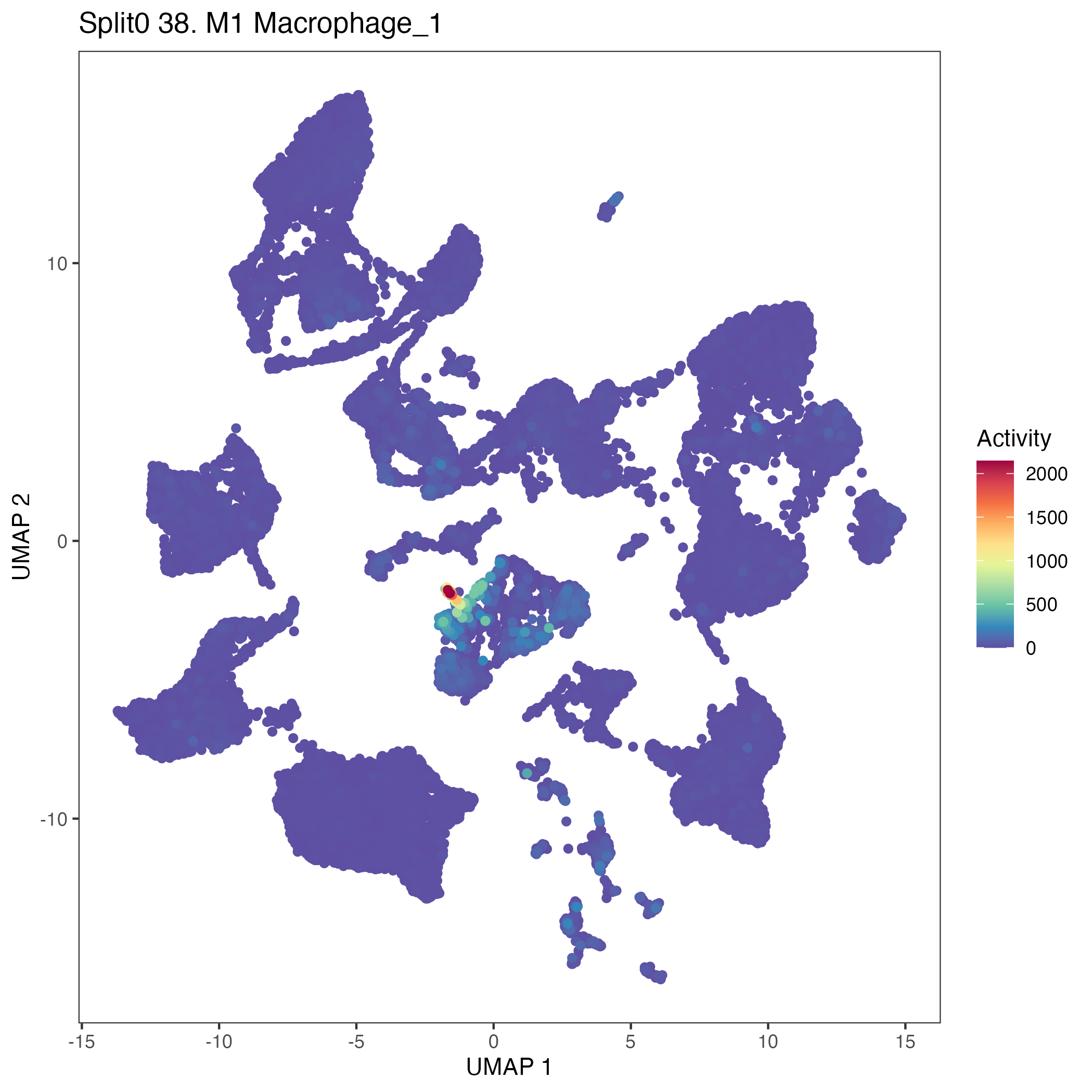
UMAP plots showing activity of gene expression program identified in GEP 38. M1 Macrophage:
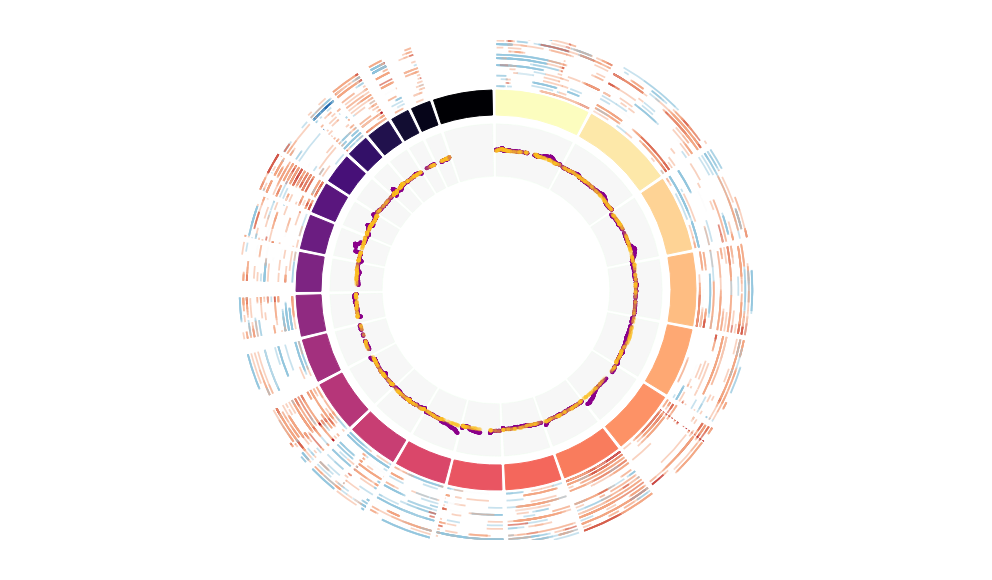
CNV Data procured from inferCNV.
Outer tracks are putative CNV regions (gains = red, losses = blue) for each patient
Inner track is expression data representing:
The top cells expressing this GEP (purple)
Random cells (n =50) from the reference set used in inferCNV (orange)
Gene set Enrichments for this program, caculated from top 50 genes
mSigDB Cell Types Gene Set:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| FAN_EMBRYONIC_CTX_MICROGLIA_2 | 1.01e-10 | 342.99 | 71.39 | 2.72e-09 | 6.81e-08 | 5IER2, IER3, FOSB, CD83, PPP1R15A |
9 |
| ZHENG_CORD_BLOOD_C5_SIMILAR_TO_HSC_C6_PUTATIVE_ALTERED_METABOLIC_STATE | 3.90e-17 | 52.23 | 24.55 | 3.73e-15 | 2.61e-14 | 13IER2, ATF3, NFKBIA, GADD45B, DUSP1, FOS, JUNB, DUSP2, NEAT1, JUN, BTG2, MAFF, NR4A1 |
97 |
| CUI_DEVELOPING_HEART_C8_MACROPHAGE | 5.61e-21 | 32.70 | 17.35 | 3.76e-18 | 3.76e-18 | 20IER3, NFKBIA, GADD45B, FOSB, DUSP1, NR4A2, CD83, IFI30, JUNB, FGL2, DUSP2, HERPUD1, NEAT1, KLF6, MS4A4A, BTG2, RILPL2, NAMPT, MAFB, C5AR1 |
275 |
| ZHENG_CORD_BLOOD_C6_HSC_MULTIPOTENT_PROGENITOR | 3.50e-12 | 35.87 | 15.49 | 1.30e-10 | 2.35e-09 | 10NFKBIA, DUSP1, NR4A2, SAT1, DUSP2, NEAT1, MAFF, RILPL2, NR4A1, NAMPT |
97 |
| AIZARANI_LIVER_C18_NK_NKT_CELLS_5 | 1.04e-12 | 32.02 | 14.40 | 4.35e-11 | 6.95e-10 | 11IER3, GADD45B, CD83, FGL2, SAT1, KLF4, TNFAIP2, RILPL2, MAFB, PHACTR1, C5AR1 |
121 |
| CUI_DEVELOPING_HEART_C5_VALVAR_CELL | 1.26e-15 | 27.75 | 13.84 | 9.36e-14 | 8.42e-13 | 15IER2, ATF3, IER3, GADD45B, EIF4A3, NR4A2, FOS, PPP1R15A, JUNB, MMP19, BTG2, SERTAD1, MAFF, NR4A1, LMNA |
208 |
| RUBENSTEIN_SKELETAL_MUSCLE_MYELOID_CELLS | 5.20e-19 | 25.49 | 13.57 | 1.16e-16 | 3.49e-16 | 20IER3, NFKBIA, FOSB, DUSP1, NR4A2, CD83, FOS, PPP1R15A, IFI30, STX11, JUNB, FGL2, SAT1, DUSP2, KLF4, HERPUD1, BTG2, NR4A1, NAMPT, C5AR1 |
347 |
| DURANTE_ADULT_OLFACTORY_NEUROEPITHELIUM_DENDRITIC_CELLS | 2.31e-11 | 29.19 | 12.68 | 7.03e-10 | 1.55e-08 | 10NFKBIA, DUSP1, IFI30, STX11, FGL2, NEAT1, TNFAIP2, NAMPT, PHACTR1, C5AR1 |
117 |
| DURANTE_ADULT_OLFACTORY_NEUROEPITHELIUM_MONOCYTES | 4.19e-07 | 40.76 | 11.90 | 5.85e-06 | 2.81e-04 | 5CD83, IFI30, FGL2, HERPUD1, PHACTR1 |
39 |
| ZHONG_PFC_MAJOR_TYPES_MICROGLIA | 2.08e-18 | 21.72 | 11.67 | 2.79e-16 | 1.40e-15 | 21IER2, IER3, NFKBIA, GADD45B, FOSB, DUSP1, NR4A2, CD83, FOS, IFI30, JUNB, SAT1, HERPUD1, NEAT1, KLF6, PLA2G7, JUN, BTG2, NR4A1, NABP1, C5AR1 |
438 |
| MANNO_MIDBRAIN_NEUROTYPES_HMGL | 7.07e-20 | 20.89 | 11.40 | 2.37e-17 | 4.74e-17 | 24IER2, ATF3, NFKBIA, GADD45B, FOSB, DUSP1, NR4A2, CD83, PPP1R15A, CD5L, SQSTM1, STX11, JUNB, FGL2, ICAM1, SAT1, DUSP2, HERPUD1, NEAT1, MS4A4A, MAFF, RILPL2, NR4A1, MAFB |
577 |
| AIZARANI_LIVER_C25_KUPFFER_CELLS_4 | 2.26e-12 | 24.36 | 11.37 | 8.92e-11 | 1.52e-09 | 12IER3, NR4A2, CD83, FGL2, SAT1, KLF4, HERPUD1, RILPL2, NAMPT, MAFB, PHACTR1, C5AR1 |
174 |
| GAO_LARGE_INTESTINE_24W_C11_PANETH_LIKE_CELL | 2.10e-15 | 20.91 | 10.80 | 1.41e-13 | 1.41e-12 | 17EIF4A3, NR4A2, CD83, IFI30, STX11, FGL2, ICAM1, DUSP2, PLA2G7, MS4A4A, TNFAIP2, ARL4C, GLA, RILPL2, NAMPT, MAFB, C5AR1 |
325 |
| TRAVAGLINI_LUNG_ADVENTITIAL_FIBROBLAST_CELL | 1.03e-14 | 21.01 | 10.69 | 6.26e-13 | 6.89e-12 | 16IER2, FOSB, DUSP1, FOS, PPP1R15A, JUNB, FGL2, KLF4, TNFAIP2, JUN, BTG2, SERTAD1, MAFF, NR4A1, TIPARP, LMNA |
296 |
| FAN_EMBRYONIC_CTX_BRAIN_MYELOID | 1.27e-10 | 24.19 | 10.57 | 3.28e-09 | 8.53e-08 | 10ATF3, NFKBIA, IFI30, FGL2, KLF4, NEAT1, TNFAIP2, NAMPT, MAFB, C5AR1 |
139 |
| LAKE_ADULT_KIDNEY_C17_COLLECTING_SYSTEM_PCS_STRESSED_DISSOC_SUBSET | 2.37e-13 | 21.51 | 10.56 | 1.13e-11 | 1.59e-10 | 14ATF3, FOSB, DUSP1, FOS, SQSTM1, SAT1, NEAT1, KLF6, ZFAND5, JUN, BTG2, NR4A1, NAMPT, LMNA |
240 |
| TRAVAGLINI_LUNG_EREG_DENDRITIC_CELL | 1.53e-18 | 19.17 | 10.43 | 2.57e-16 | 1.03e-15 | 23ATF3, IER3, GADD45B, FOSB, DUSP1, NR4A2, CD83, FOS, IFI30, JDP2, FGL2, ICAM1, SAT1, KLF4, NEAT1, ZFAND5, MS4A4A, MMP19, RILPL2, NAMPT, SLC1A3, MAFB, C5AR1 |
579 |
| CUI_DEVELOPING_HEART_VASCULAR_ENDOTHELIAL_CELL | 7.60e-12 | 21.80 | 10.20 | 2.55e-10 | 5.10e-09 | 12IER2, NFKBIA, GADD45B, FOSB, PPP1R15A, JUNB, FGL2, VCAM1, ICAM1, KLF4, JUN, BTG2 |
193 |
| DURANTE_ADULT_OLFACTORY_NEUROEPITHELIUM_MACROPHAGES | 2.74e-08 | 27.47 | 10.09 | 4.60e-07 | 1.84e-05 | 7DUSP1, NR4A2, CD83, IFI30, FGL2, HERPUD1, PHACTR1 |
81 |
| TRAVAGLINI_LUNG_IGSF21_DENDRITIC_CELL | 9.74e-07 | 33.82 | 9.97 | 1.26e-05 | 6.53e-04 | 5ATF3, CD83, MS4A4A, LMNA, MAFB |
46 |
Dowload full table
mSigDB Hallmark Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 1.50e-32 | 71.50 | 38.46 | 7.51e-31 | 7.51e-31 | 25IER2, ATF3, IER3, NFKBIA, GADD45B, FOSB, DUSP1, NR4A2, CD83, FOS, PPP1R15A, SQSTM1, JUNB, ICAM1, SAT1, DUSP2, KLF4, KLF6, TNFAIP2, JUN, BTG2, MAFF, NR4A1, NAMPT, TIPARP |
200 |
| HALLMARK_UV_RESPONSE_UP | 9.16e-09 | 18.41 | 7.73 | 2.29e-07 | 4.58e-07 | 9ATF3, NFKBIA, FOSB, FOS, SQSTM1, JUNB, ICAM1, BTG2, NR4A1 |
158 |
| HALLMARK_APOPTOSIS | 1.90e-07 | 15.56 | 6.20 | 1.90e-06 | 9.52e-06 | 8ATF3, IER3, GADD45B, SQSTM1, SAT1, JUN, BTG2, LMNA |
161 |
| HALLMARK_HYPOXIA | 6.91e-08 | 14.37 | 6.05 | 8.63e-07 | 3.45e-06 | 9ATF3, IER3, DUSP1, FOS, PPP1R15A, KLF6, JUN, MAFF, TIPARP |
200 |
| HALLMARK_P53_PATHWAY | 6.91e-08 | 14.37 | 6.05 | 8.63e-07 | 3.45e-06 | 9ATF3, IER3, FOS, PPP1R15A, IFI30, SAT1, KLF4, JUN, BTG2 |
200 |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | 1.19e-05 | 10.54 | 3.95 | 8.48e-05 | 5.94e-04 | 7NFKBIA, IFI30, FGL2, VCAM1, ICAM1, TNFAIP2, NAMPT |
200 |
| HALLMARK_MTORC1_SIGNALING | 1.19e-05 | 10.54 | 3.95 | 8.48e-05 | 5.94e-04 | 7PPP1R15A, SQSTM1, IFI30, FGL2, GLA, BTG2, NAMPT |
200 |
| HALLMARK_IL2_STAT5_SIGNALING | 1.20e-04 | 8.83 | 3.04 | 6.87e-04 | 6.02e-03 | 6GADD45B, CD83, FGL2, KLF6, MAFF, SMPDL3A |
199 |
| HALLMARK_INFLAMMATORY_RESPONSE | 1.24e-04 | 8.78 | 3.02 | 6.87e-04 | 6.18e-03 | 6NFKBIA, ICAM1, KLF6, BTG2, NAMPT, C5AR1 |
200 |
| HALLMARK_UNFOLDED_PROTEIN_RESPONSE | 1.01e-03 | 9.97 | 2.56 | 5.05e-03 | 5.05e-02 | 4ATF3, EIF4A3, HERPUD1, NABP1 |
113 |
| HALLMARK_ESTROGEN_RESPONSE_EARLY | 7.71e-03 | 5.55 | 1.44 | 2.96e-02 | 3.85e-01 | 4FOS, KLF4, GLA, TIPARP |
200 |
| HALLMARK_ESTROGEN_RESPONSE_LATE | 7.71e-03 | 5.55 | 1.44 | 2.96e-02 | 3.85e-01 | 4FOS, DUSP2, KLF4, GLA |
200 |
| HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | 7.71e-03 | 5.55 | 1.44 | 2.96e-02 | 3.85e-01 | 4GADD45B, VCAM1, SAT1, JUN |
200 |
| HALLMARK_TGF_BETA_SIGNALING | 1.94e-02 | 10.01 | 1.15 | 6.91e-02 | 9.68e-01 | 2PPP1R15A, JUNB |
54 |
| HALLMARK_KRAS_SIGNALING_UP | 4.35e-02 | 4.05 | 0.80 | 1.45e-01 | 1.00e+00 | 3PPP1R15A, KLF4, MAFB |
200 |
| HALLMARK_ADIPOGENESIS | 1.85e-01 | 2.63 | 0.31 | 4.40e-01 | 1.00e+00 | 2NABP1, PPP1R15B |
200 |
| HALLMARK_APICAL_JUNCTION | 1.85e-01 | 2.63 | 0.31 | 4.40e-01 | 1.00e+00 | 2VCAM1, ICAM1 |
200 |
| HALLMARK_COMPLEMENT | 1.85e-01 | 2.63 | 0.31 | 4.40e-01 | 1.00e+00 | 2PLA2G7, MAFF |
200 |
| HALLMARK_KRAS_SIGNALING_DN | 1.85e-01 | 2.63 | 0.31 | 4.40e-01 | 1.00e+00 | 2NR4A2, BTG2 |
200 |
| HALLMARK_PANCREAS_BETA_CELLS | 1.47e-01 | 6.54 | 0.16 | 4.40e-01 | 1.00e+00 | 1MAFB |
40 |
Dowload full table
KEGG Pathways:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| KEGG_MAPK_SIGNALING_PATHWAY | 5.69e-04 | 6.53 | 2.25 | 1.06e-01 | 1.06e-01 | 6GADD45B, DUSP1, FOS, DUSP2, JUN, NR4A1 |
267 |
| KEGG_LEISHMANIA_INFECTION | 2.88e-03 | 11.56 | 2.25 | 2.00e-01 | 5.36e-01 | 3NFKBIA, FOS, JUN |
72 |
| KEGG_B_CELL_RECEPTOR_SIGNALING_PATHWAY | 3.23e-03 | 11.08 | 2.16 | 2.00e-01 | 6.01e-01 | 3NFKBIA, FOS, JUN |
75 |
| KEGG_TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY | 7.58e-03 | 8.06 | 1.58 | 3.29e-01 | 1.00e+00 | 3NFKBIA, FOS, JUN |
102 |
| KEGG_T_CELL_RECEPTOR_SIGNALING_PATHWAY | 8.85e-03 | 7.60 | 1.49 | 3.29e-01 | 1.00e+00 | 3NFKBIA, FOS, JUN |
108 |
| KEGG_COLORECTAL_CANCER | 2.50e-02 | 8.68 | 1.00 | 7.76e-01 | 1.00e+00 | 2FOS, JUN |
62 |
| KEGG_EPITHELIAL_CELL_SIGNALING_IN_HELICOBACTER_PYLORI_INFECTION | 2.97e-02 | 7.89 | 0.91 | 7.89e-01 | 1.00e+00 | 2NFKBIA, JUN |
68 |
| KEGG_LEUKOCYTE_TRANSENDOTHELIAL_MIGRATION | 7.67e-02 | 4.57 | 0.53 | 1.00e+00 | 1.00e+00 | 2VCAM1, ICAM1 |
116 |
| KEGG_PATHWAYS_IN_CANCER | 1.32e-01 | 2.48 | 0.49 | 1.00e+00 | 1.00e+00 | 3NFKBIA, FOS, JUN |
325 |
| KEGG_NEUROTROPHIN_SIGNALING_PATHWAY | 8.82e-02 | 4.20 | 0.49 | 1.00e+00 | 1.00e+00 | 2NFKBIA, JUN |
126 |
| KEGG_CELL_ADHESION_MOLECULES_CAMS | 9.66e-02 | 3.98 | 0.46 | 1.00e+00 | 1.00e+00 | 2VCAM1, ICAM1 |
133 |
| KEGG_GLYCOSPHINGOLIPID_BIOSYNTHESIS_GLOBO_SERIES | 5.43e-02 | 19.60 | 0.45 | 1.00e+00 | 1.00e+00 | 1GLA |
14 |
| KEGG_NICOTINATE_AND_NICOTINAMIDE_METABOLISM | 9.13e-02 | 11.08 | 0.26 | 1.00e+00 | 1.00e+00 | 1NAMPT |
24 |
| KEGG_GALACTOSE_METABOLISM | 9.85e-02 | 10.19 | 0.24 | 1.00e+00 | 1.00e+00 | 1GLA |
26 |
| KEGG_ETHER_LIPID_METABOLISM | 1.23e-01 | 7.97 | 0.19 | 1.00e+00 | 1.00e+00 | 1PLA2G7 |
33 |
| KEGG_SNARE_INTERACTIONS_IN_VESICULAR_TRANSPORT | 1.41e-01 | 6.89 | 0.17 | 1.00e+00 | 1.00e+00 | 1STX11 |
38 |
| KEGG_SPHINGOLIPID_METABOLISM | 1.44e-01 | 6.71 | 0.16 | 1.00e+00 | 1.00e+00 | 1GLA |
39 |
| KEGG_GLYCEROLIPID_METABOLISM | 1.77e-01 | 5.31 | 0.13 | 1.00e+00 | 1.00e+00 | 1GLA |
49 |
| KEGG_ARGININE_AND_PROLINE_METABOLISM | 1.94e-01 | 4.81 | 0.12 | 1.00e+00 | 1.00e+00 | 1SAT1 |
54 |
| KEGG_CYTOSOLIC_DNA_SENSING_PATHWAY | 1.97e-01 | 4.72 | 0.12 | 1.00e+00 | 1.00e+00 | 1NFKBIA |
55 |
Dowload full table
CHR Positional Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| chr1q32 | 8.51e-02 | 3.03 | 0.60 | 1.00e+00 | 1.00e+00 | 3ATF3, BTG2, PPP1R15B |
266 |
| chr19p13 | 2.13e-01 | 1.81 | 0.56 | 1.00e+00 | 1.00e+00 | 5IER2, GADD45B, IFI30, JUNB, ICAM1 |
773 |
| chr6p23 | 5.81e-02 | 18.20 | 0.42 | 1.00e+00 | 1.00e+00 | 1CD83 |
15 |
| chr14q24 | 1.72e-01 | 2.76 | 0.32 | 1.00e+00 | 1.00e+00 | 2FOS, JDP2 |
191 |
| chr5q35 | 2.16e-01 | 2.37 | 0.28 | 1.00e+00 | 1.00e+00 | 2DUSP1, SQSTM1 |
222 |
| chr20q12 | 9.13e-02 | 11.08 | 0.26 | 1.00e+00 | 1.00e+00 | 1MAFB |
24 |
| chr19q13 | 1.00e+00 | 0.94 | 0.24 | 1.00e+00 | 1.00e+00 | 4FOSB, PPP1R15A, SERTAD1, C5AR1 |
1165 |
| chr16q13 | 1.47e-01 | 6.54 | 0.16 | 1.00e+00 | 1.00e+00 | 1HERPUD1 |
40 |
| chr12q13 | 6.71e-01 | 1.29 | 0.15 | 1.00e+00 | 1.00e+00 | 2MMP19, NR4A1 |
407 |
| chr6p24 | 2.03e-01 | 4.55 | 0.11 | 1.00e+00 | 1.00e+00 | 1PHACTR1 |
57 |
| chr14q13 | 2.09e-01 | 4.40 | 0.11 | 1.00e+00 | 1.00e+00 | 1NFKBIA |
59 |
| chr1q22 | 2.43e-01 | 3.70 | 0.09 | 1.00e+00 | 1.00e+00 | 1LMNA |
70 |
| chr6q24 | 2.49e-01 | 3.59 | 0.09 | 1.00e+00 | 1.00e+00 | 1STX11 |
72 |
| chr10p15 | 2.90e-01 | 3.00 | 0.07 | 1.00e+00 | 1.00e+00 | 1KLF6 |
86 |
| chr1p21 | 3.25e-01 | 2.60 | 0.06 | 1.00e+00 | 1.00e+00 | 1VCAM1 |
99 |
| chr2q32 | 3.49e-01 | 2.38 | 0.06 | 1.00e+00 | 1.00e+00 | 1NABP1 |
108 |
| chr6q22 | 3.77e-01 | 2.16 | 0.05 | 1.00e+00 | 1.00e+00 | 1SMPDL3A |
119 |
| chr2q24 | 3.91e-01 | 2.06 | 0.05 | 1.00e+00 | 1.00e+00 | 1NR4A2 |
125 |
| chr5p13 | 3.99e-01 | 2.01 | 0.05 | 1.00e+00 | 1.00e+00 | 1SLC1A3 |
128 |
| chr9q31 | 3.99e-01 | 2.01 | 0.05 | 1.00e+00 | 1.00e+00 | 1KLF4 |
128 |
Dowload full table
Transcription Factor Targets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| SRF_01 | 4.78e-05 | 23.08 | 5.81 | 5.40e-03 | 5.41e-02 | 4IER2, FOSB, FOS, JUNB |
51 |
| CREB_Q2 | 8.15e-07 | 10.56 | 4.46 | 4.62e-04 | 9.24e-04 | 9ATF3, GADD45B, FOSB, DUSP1, NR4A2, FOS, PPP1R15A, HERPUD1, MAFF |
269 |
| GTF2A2_TARGET_GENES | 5.18e-08 | 8.63 | 4.18 | 5.86e-05 | 5.86e-05 | 13ATF3, IER3, GADD45B, FOSB, DUSP1, FOS, PPP1R15A, SQSTM1, JUNB, SAT1, HERPUD1, NEAT1, KLF6 |
522 |
| YWATTWNNRGCT_UNKNOWN | 1.74e-04 | 16.21 | 4.12 | 1.41e-02 | 1.97e-01 | 4ATF3, NFKBIA, ARL4C, NR4A1 |
71 |
| PSMB5_TARGET_GENES | 2.38e-06 | 9.21 | 3.90 | 8.99e-04 | 2.70e-03 | 9ATF3, IER3, DUSP1, PPP1R15A, JUNB, BTG2, LMNA, NABP1, PPP1R15B |
307 |
| ATF_01 | 8.04e-06 | 9.19 | 3.69 | 1.82e-03 | 9.11e-03 | 8ATF3, FOSB, DUSP1, NR4A2, FOS, PPP1R15A, ZFAND5, MAFF |
267 |
| SRF_C | 1.88e-05 | 9.78 | 3.67 | 3.56e-03 | 2.13e-02 | 7IER2, FOSB, FOS, JUNB, DUSP2, KLF6, NR4A1 |
215 |
| CREB_Q2_01 | 2.58e-05 | 9.29 | 3.49 | 4.09e-03 | 2.93e-02 | 7FOSB, DUSP1, NR4A2, PPP1R15A, ZFAND5, JUN, MAFF |
226 |
| SRF_Q4 | 2.89e-05 | 9.12 | 3.43 | 4.09e-03 | 3.27e-02 | 7IER2, FOSB, FOS, JUNB, DUSP2, KLF6, NR4A1 |
230 |
| CCAWWNAAGG_SRF_Q4 | 3.96e-04 | 12.93 | 3.31 | 2.49e-02 | 4.49e-01 | 4FOSB, DUSP2, KLF6, NR4A1 |
88 |
| CEBPDELTA_Q6 | 4.51e-05 | 8.48 | 3.19 | 5.40e-03 | 5.11e-02 | 7ATF3, FOSB, NR4A2, FOS, JUN, GLA, NR4A1 |
247 |
| ATF3_Q6 | 5.24e-05 | 8.27 | 3.11 | 5.40e-03 | 5.94e-02 | 7ATF3, FOSB, DUSP1, PPP1R15A, ZFAND5, JUN, MAFF |
253 |
| CREB_Q4 | 8.20e-05 | 7.68 | 2.89 | 7.74e-03 | 9.29e-02 | 7ATF3, FOSB, DUSP1, NR4A2, FOS, PPP1R15A, MAFF |
272 |
| STAT3_01 | 3.40e-03 | 26.01 | 2.87 | 1.10e-01 | 1.00e+00 | 2ICAM1, MAFF |
22 |
| CREB_Q4_01 | 1.92e-04 | 8.08 | 2.78 | 1.45e-02 | 2.17e-01 | 6FOSB, DUSP1, NR4A2, PPP1R15A, ZFAND5, MAFF |
217 |
| SRF_Q5_01 | 2.32e-04 | 7.78 | 2.68 | 1.65e-02 | 2.63e-01 | 6IER2, ATF3, FOSB, FOS, JUNB, KLF6 |
225 |
| CHAF1B_TARGET_GENES | 8.03e-06 | 5.03 | 2.50 | 1.82e-03 | 9.10e-03 | 14IER3, NFKBIA, GADD45B, DUSP1, PPP1R15A, SQSTM1, JUNB, ICAM1, SAT1, DUSP2, HERPUD1, KLF6, TIPARP, NABP1 |
981 |
| SRF_Q6 | 3.88e-04 | 7.04 | 2.43 | 2.49e-02 | 4.40e-01 | 6FOSB, FOS, JUNB, DUSP2, KLF6, NR4A1 |
248 |
| GTF2E2_TARGET_GENES | 1.59e-04 | 5.91 | 2.38 | 1.39e-02 | 1.81e-01 | 8ATF3, GADD45B, DUSP1, PPP1R15A, JUNB, HERPUD1, NEAT1, KLF6 |
411 |
| STAT5B_01 | 4.49e-04 | 6.85 | 2.36 | 2.56e-02 | 5.08e-01 | 6NFKBIA, NR4A2, FOS, ICAM1, KLF4, MAFF |
255 |
Dowload full table
GO Biological Processes:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GOBP_MEMBRANE_TO_MEMBRANE_DOCKING | 1.54e-04 | 172.68 | 14.09 | 7.69e-02 | 1.00e+00 | 2VCAM1, ICAM1 |
5 |
| GOBP_RESPONSE_TO_CORTICOTROPIN_RELEASING_HORMONE | 1.54e-04 | 172.68 | 14.09 | 7.69e-02 | 1.00e+00 | 2NR4A2, NR4A1 |
5 |
| GOBP_NEGATIVE_REGULATION_OF_PERK_MEDIATED_UNFOLDED_PROTEIN_RESPONSE | 2.31e-04 | 129.66 | 11.45 | 1.08e-01 | 1.00e+00 | 2PPP1R15A, PPP1R15B |
6 |
| GOBP_INTEGRATED_STRESS_RESPONSE_SIGNALING | 3.63e-06 | 47.11 | 11.40 | 1.36e-02 | 2.71e-02 | 4ATF3, PPP1R15A, HERPUD1, PPP1R15B |
27 |
| GOBP_PEPTIDYL_SERINE_DEPHOSPHORYLATION | 5.49e-05 | 49.70 | 8.99 | 5.20e-02 | 4.11e-01 | 3DUSP1, PPP1R15A, PPP1R15B |
19 |
| GOBP_REGULATION_OF_TRANSLATION_INITIATION_IN_RESPONSE_TO_ENDOPLASMIC_RETICULUM_STRESS | 4.28e-04 | 86.37 | 8.33 | 1.60e-01 | 1.00e+00 | 2PPP1R15A, PPP1R15B |
8 |
| GOBP_T_CELL_ACTIVATION_VIA_T_CELL_RECEPTOR_CONTACT_WITH_ANTIGEN_BOUND_TO_MHC_MOLECULE_ON_ANTIGEN_PRESENTING_CELL | 6.85e-04 | 64.86 | 6.55 | 2.08e-01 | 1.00e+00 | 2FGL2, ICAM1 |
10 |
| GOBP_REGULATION_OF_TRANSLATION_IN_RESPONSE_TO_ENDOPLASMIC_RETICULUM_STRESS | 8.35e-04 | 57.69 | 5.91 | 2.15e-01 | 1.00e+00 | 2PPP1R15A, PPP1R15B |
11 |
| GOBP_REGULATION_OF_PERK_MEDIATED_UNFOLDED_PROTEIN_RESPONSE | 8.35e-04 | 57.69 | 5.91 | 2.15e-01 | 1.00e+00 | 2PPP1R15A, PPP1R15B |
11 |
| GOBP_ER_NUCLEUS_SIGNALING_PATHWAY | 5.56e-05 | 22.15 | 5.58 | 5.20e-02 | 4.16e-01 | 4ATF3, PPP1R15A, HERPUD1, PPP1R15B |
53 |
| GOBP_NEGATIVE_REGULATION_OF_MUSCLE_ADAPTATION | 1.18e-03 | 47.19 | 4.96 | 2.43e-01 | 1.00e+00 | 2KLF4, LMNA |
13 |
| GOBP_RESPONSE_TO_CAMP | 3.15e-05 | 15.77 | 4.77 | 3.93e-02 | 2.36e-01 | 5FOSB, DUSP1, FOS, MMP19, JUN |
93 |
| GOBP_SKELETAL_MUSCLE_CELL_DIFFERENTIATION | 1.17e-04 | 18.10 | 4.59 | 7.69e-02 | 8.72e-01 | 4ATF3, FOS, BTG2, MAFF |
64 |
| GOBP_NEGATIVE_REGULATION_OF_ENDOPLASMIC_RETICULUM_UNFOLDED_PROTEIN_RESPONSE | 1.37e-03 | 43.30 | 4.59 | 2.50e-01 | 1.00e+00 | 2PPP1R15A, PPP1R15B |
14 |
| GOBP_REGULATION_OF_TRANSLATIONAL_INITIATION_IN_RESPONSE_TO_STRESS | 1.58e-03 | 39.95 | 4.27 | 2.74e-01 | 1.00e+00 | 2PPP1R15A, PPP1R15B |
15 |
| GOBP_POSITIVE_REGULATION_OF_PRI_MIRNA_TRANSCRIPTION_BY_RNA_POLYMERASE_II | 5.25e-04 | 21.53 | 4.11 | 1.79e-01 | 1.00e+00 | 3FOS, KLF4, JUN |
40 |
| GOBP_NEGATIVE_REGULATION_OF_RESPONSE_TO_ENDOPLASMIC_RETICULUM_STRESS | 6.96e-04 | 19.44 | 3.72 | 2.08e-01 | 1.00e+00 | 3PPP1R15A, HERPUD1, PPP1R15B |
44 |
| GOBP_ENDOPLASMIC_RETICULUM_UNFOLDED_PROTEIN_RESPONSE | 1.38e-04 | 11.38 | 3.47 | 7.69e-02 | 1.00e+00 | 5ATF3, PPP1R15A, HERPUD1, LMNA, PPP1R15B |
127 |
| GOBP_RESPONSE_TO_ORGANOPHOSPHORUS | 1.54e-04 | 11.10 | 3.38 | 7.69e-02 | 1.00e+00 | 5FOSB, DUSP1, FOS, MMP19, JUN |
130 |
| GOBP_RESPONSE_TO_CORTICOSTERONE | 2.81e-03 | 28.89 | 3.17 | 3.63e-01 | 1.00e+00 | 2FOSB, FOS |
20 |
Dowload full table
Immunological Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GSE14769_UNSTIM_VS_40MIN_LPS_BMDM_DN | 4.34e-22 | 42.31 | 22.14 | 2.12e-18 | 2.12e-18 | 19IER2, ATF3, IER3, NFKBIA, GADD45B, DUSP1, FOS, PPP1R15A, SQSTM1, JUNB, DUSP2, HERPUD1, KLF6, ZFAND5, JUN, BTG2, MAFF, TIPARP, NABP1 |
200 |
| GSE14769_UNSTIM_VS_80MIN_LPS_BMDM_DN | 1.62e-20 | 38.83 | 20.12 | 2.89e-17 | 7.91e-17 | 18IER2, ATF3, GADD45B, DUSP1, CD83, FOS, SQSTM1, STX11, ICAM1, HERPUD1, KLF6, ZFAND5, TNFAIP2, BTG2, MAFF, NAMPT, TIPARP, NABP1 |
199 |
| GSE14769_UNSTIM_VS_60MIN_LPS_BMDM_DN | 1.78e-20 | 38.62 | 20.01 | 2.89e-17 | 8.66e-17 | 18IER2, ATF3, IER3, GADD45B, DUSP1, CD83, FOS, SQSTM1, STX11, ICAM1, DUSP2, HERPUD1, KLF6, ZFAND5, TNFAIP2, JUN, BTG2, TIPARP |
200 |
| GSE19923_WT_VS_HEB_AND_E2A_KO_DP_THYMOCYTE_DN | 6.65e-19 | 35.18 | 18.04 | 8.10e-16 | 3.24e-15 | 17IER2, ATF3, IER3, NFKBIA, DUSP1, FOS, PPP1R15A, JUNB, KLF6, ZFAND5, BTG2, SERTAD1, NR4A1, NAMPT, TIPARP, NABP1, PPP1R15B |
200 |
| GSE36891_POLYIC_TLR3_VS_PAM_TLR2_STIM_PERITONEAL_MACROPHAGE_UP | 4.78e-16 | 34.95 | 17.02 | 3.81e-13 | 2.33e-12 | 14IER2, ATF3, IER3, FOSB, DUSP1, NR4A2, FOS, JUNB, DUSP2, KLF4, JUN, BTG2, MAFF, NR4A1 |
153 |
| GSE29617_CTRL_VS_DAY7_TIV_FLU_VACCINE_PBMC_2008_UP | 1.64e-17 | 32.66 | 16.52 | 1.60e-14 | 8.01e-14 | 16ATF3, FOSB, DUSP1, CD83, FOS, STX11, JUNB, SAT1, DUSP2, ZFAND5, BTG2, NR4A1, NAMPT, TIPARP, PHACTR1, C5AR1 |
196 |
| GSE37605_TREG_VS_TCONV_NOD_FOXP3_FUSION_GFP_UP | 1.25e-14 | 32.04 | 15.27 | 5.09e-12 | 6.11e-11 | 13IER2, ATF3, IER3, FOSB, DUSP1, NR4A2, FOS, PPP1R15A, JUNB, JUN, BTG2, MAFF, NR4A1 |
150 |
| GSE23925_LIGHT_ZONE_VS_NAIVE_BCELL_UP | 6.53e-16 | 29.09 | 14.50 | 3.81e-13 | 3.18e-12 | 15IER2, ATF3, FOSB, DUSP1, NR4A2, FOS, PPP1R15A, JUNB, DUSP2, NEAT1, MAFF, RILPL2, NR4A1, NABP1, PPP1R15B |
199 |
| GSE42021_TREG_PLN_VS_CD24HI_TREG_THYMUS_UP | 7.03e-16 | 28.93 | 14.43 | 3.81e-13 | 3.43e-12 | 15IER2, IER3, NFKBIA, GADD45B, DUSP1, CD83, FOS, PPP1R15A, SQSTM1, JUNB, ICAM1, DUSP2, TNFAIP2, BTG2, NR4A1 |
200 |
| GSE27434_WT_VS_DNMT1_KO_TREG_DN | 7.03e-16 | 28.93 | 14.43 | 3.81e-13 | 3.43e-12 | 15IER2, NFKBIA, GADD45B, DUSP1, CD83, FOS, PPP1R15A, SQSTM1, JUNB, ICAM1, DUSP2, TNFAIP2, BTG2, SERTAD1, NR4A1 |
200 |
| GSE2706_UNSTIM_VS_2H_LPS_AND_R848_DC_DN | 1.05e-14 | 27.47 | 13.43 | 4.65e-12 | 5.11e-11 | 14ATF3, NFKBIA, DUSP1, NR4A2, PPP1R15A, STX11, JUNB, ICAM1, DUSP2, TNFAIP2, BTG2, RILPL2, NAMPT, NABP1 |
191 |
| GSE37605_C57BL6_VS_NOD_FOXP3_FUSION_GFP_TREG_DN | 1.05e-14 | 27.47 | 13.43 | 4.65e-12 | 5.11e-11 | 14IER2, ATF3, IER3, FOSB, DUSP1, FOS, JUNB, JDP2, KLF4, ZFAND5, JUN, BTG2, MAFF, NR4A1 |
191 |
| GSE9988_LOW_LPS_VS_CTRL_TREATED_MONOCYTE_UP | 1.50e-14 | 26.70 | 13.07 | 5.61e-12 | 7.30e-11 | 14IER3, NFKBIA, GADD45B, DUSP1, NR4A2, CD83, PPP1R15A, ICAM1, DUSP2, JUN, BTG2, MAFF, NR4A1, PPP1R15B |
196 |
| GSE45365_NK_CELL_VS_CD11B_DC_DN | 1.85e-14 | 26.27 | 12.86 | 6.42e-12 | 8.99e-11 | 14IER3, NFKBIA, DUSP1, FOS, STX11, JUNB, JDP2, ICAM1, TNFAIP2, BTG2, MAFF, TIPARP, MAFB, PPP1R15B |
199 |
| GSE26343_UNSTIM_VS_LPS_STIM_NFAT5_KO_MACROPHAGE_DN | 1.98e-14 | 26.13 | 12.79 | 6.42e-12 | 9.64e-11 | 14IER2, IER3, GADD45B, DUSP1, NR4A2, CD83, JUNB, ICAM1, DUSP2, HERPUD1, TNFAIP2, BTG2, MAFF, NR4A1 |
200 |
| GSE37605_FOXP3_FUSION_GFP_VS_IRES_GFP_TREG_C57BL6_UP | 1.87e-13 | 25.52 | 12.23 | 5.68e-11 | 9.09e-10 | 13ATF3, IER3, FOSB, DUSP1, NR4A2, FOS, PPP1R15A, JUNB, JDP2, JUN, BTG2, MAFF, NR4A1 |
185 |
| GSE36891_UNSTIM_VS_POLYIC_TLR3_STIM_PERITONEAL_MACROPHAGE_UP | 1.01e-11 | 25.53 | 11.55 | 1.97e-09 | 4.92e-08 | 11IER2, ATF3, IER3, FOSB, NR4A2, FOS, PPP1R15A, TNFAIP2, BTG2, MAFF, NR4A1 |
149 |
| GSE9988_LPS_VS_VEHICLE_TREATED_MONOCYTE_UP | 3.89e-13 | 23.98 | 11.50 | 1.06e-10 | 1.90e-09 | 13IER3, NFKBIA, GADD45B, DUSP1, CD83, PPP1R15A, ICAM1, DUSP2, TNFAIP2, JUN, BTG2, MAFF, PPP1R15B |
196 |
| GSE45365_NK_CELL_VS_BCELL_UP | 4.43e-13 | 23.72 | 11.38 | 1.06e-10 | 2.16e-09 | 13IER2, ATF3, FOSB, DUSP1, CD83, FOS, JUNB, JDP2, FGL2, VCAM1, ICAM1, NABP1, PPP1R15B |
198 |
| GSE17721_CTRL_VS_LPS_1H_BMDC_DN | 4.72e-13 | 23.60 | 11.32 | 1.06e-10 | 2.30e-09 | 13IER2, ATF3, IER3, FOS, PPP1R15A, ICAM1, DUSP2, TNFAIP2, JUN, MAFF, NR4A1, TIPARP, NABP1 |
199 |
Top Ranked Transcription Factors for this Gene Expression Program:
| Gene Symbol | TF Rank | DNA Binding Domain | Motif Status | IUPAC PWM | GTEx | DepMap | Decartes |
|---|---|---|---|---|---|---|---|
| ATF3 | 2 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| NFKBIA | 4 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | This is I kappa B alpha, which interacts with NFkappaB. No DBD, and evidence indicates that it is not a TF. |
| FOSB | 7 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| NR4A2 | 9 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| FOS | 11 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| JUNB | 17 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | Prefers forming heterodimers with FOS; FOSB; FOSL1 and FOSL2 over homodimers (PMID:12805554); but, clearly can bind DNA specifically in vitro. |
| JDP2 | 18 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| ICAM1 | 21 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| KLF4 | 24 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| KLF6 | 27 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| ZFAND5 | 29 | No | ssDNA/RNA binding | Not a DNA binding protein | No motif | None | An RNA-binding protein that recognises 3UTR located AU-rich elements in mRNA molecules to stabilize them (PMID: 22665488) |
| JUN | 34 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| GLA | 35 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | GLA appears to lack a functional SAND domain. It encodes a well-characterized enzyme, Galactosidase Alpha. Most of the polypeptide encodes a glycoside hydrolase domain and most of the rest encodes another pfam domain called Glycoside hydrolase family 27. It has a low-scoring SAND domain match that almost certainly overlaps with a known domain related to its function as an enzyme. |
| BTG2 | 36 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Transcriptional co-factor (PMID: 10617598) |
| SERTAD1 | 37 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Transcriptional co-factor; no evidence for DNA-binding activity |
| MAFF | 38 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| NR4A1 | 40 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| TIPARP | 43 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| MAFB | 46 | Yes | Known motif | Monomer or homomultimer | 100 perc ID - in vitro | None | PDB:2WTY is a homodimer crystallised with TAATTGCTGACTCAGCAAAT sequence |
| SGK1 | 51 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
QQ Plot showing correlations with other GEPs in this dataset, calculated by Spearman correlation:
Interactive QQ-plot of gene loadings:
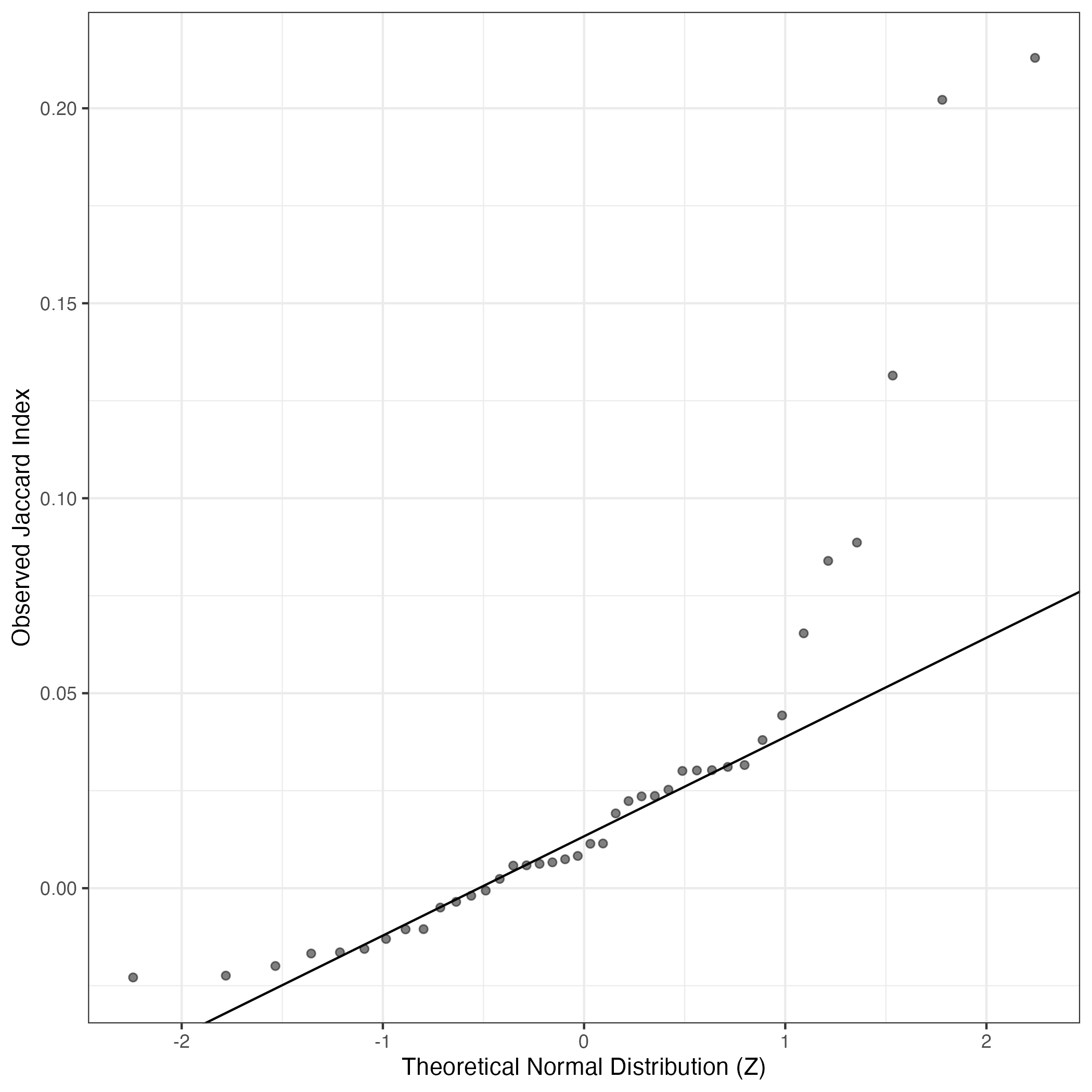
A similar QQ-plot as above, but only for instances where the H value is e.g. > 25, i.e. we are confident that the expression program is active above noise. Agreemenet between these binary vectors is tested using the Jaccard Index, with the P-values calculated by an exact test:
Interactive QQ-plot:
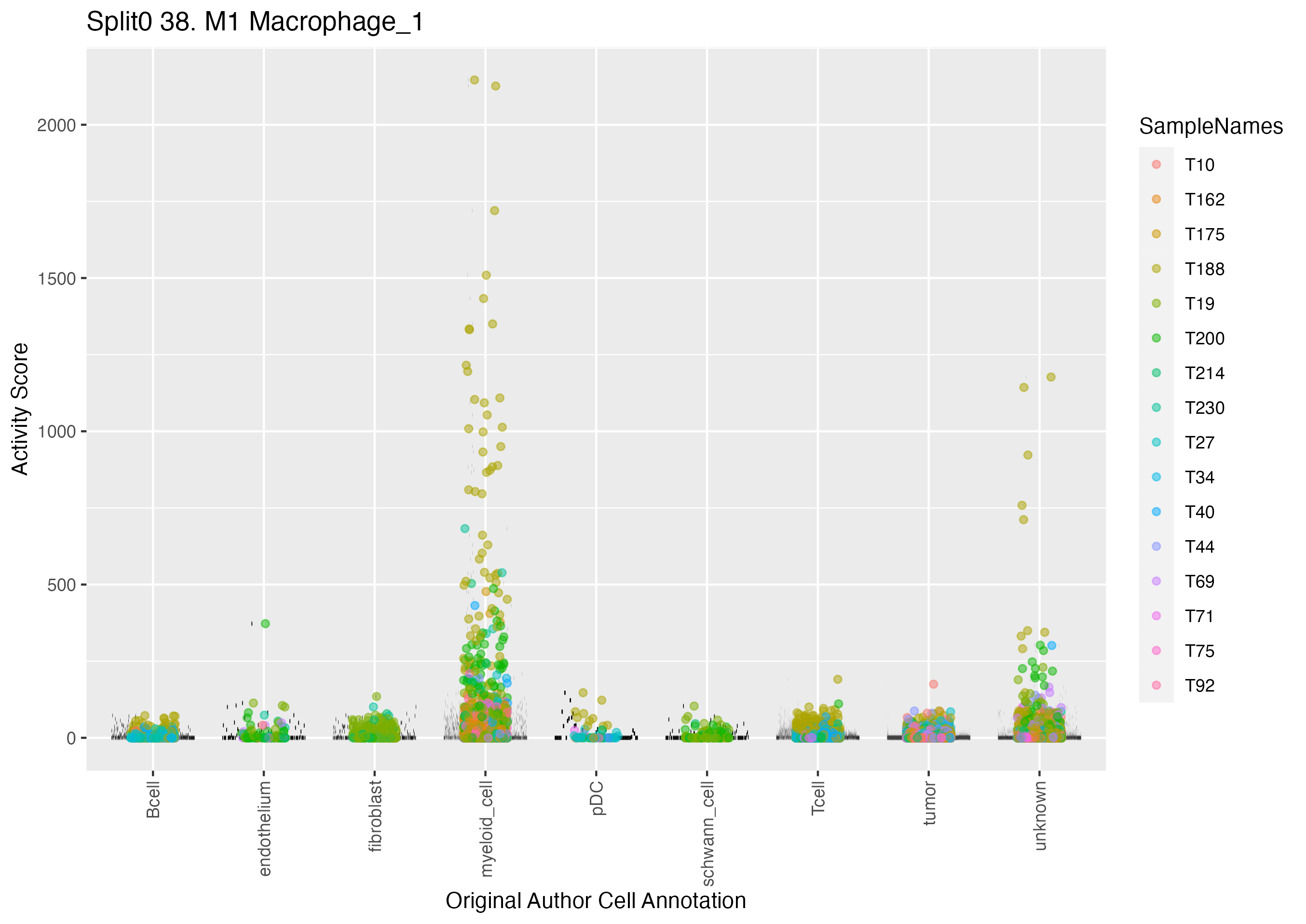
Singler cell type annotations for the top 50 cells on this program.
| Cell ID | Singler label | Singler Delta | Activity Score | Top Singler Raw Scores |
|---|---|---|---|---|
| T188_AGAAATGCAACAAGAT-1 | Macrophage:monocyte-derived:IL-4/Dex/cntrl | 0.18 | 2146.07 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.4, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.39, DC:monocyte-derived:AEC-conditioned: 0.39, Macrophage:monocyte-derived:M-CSF: 0.39, Macrophage:monocyte-derived:M-CSF/IFNg: 0.39, DC:monocyte-derived:LPS: 0.39, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.39, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.39, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.39, DC:monocyte-derived:Galectin-1: 0.39 |
| T188_AGCATCATCGCTACAA-1 | Macrophage:monocyte-derived | 0.17 | 2126.38 | Raw ScoresMacrophage:monocyte-derived:IL-4/Dex/cntrl: 0.42, Macrophage:monocyte-derived:M-CSF: 0.42, DC:monocyte-derived:anti-DC-SIGN_2h: 0.41, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.41, DC:monocyte-derived:antiCD40/VAF347: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.41, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.41, DC:monocyte-derived: 0.41, Macrophage:monocyte-derived:M-CSF/Pam3Cys: 0.4, DC:monocyte-derived:AEC-conditioned: 0.4 |
| T188_TCAAGACTCGCAGATT-1 | Macrophage:monocyte-derived:M-CSF | 0.19 | 1719.91 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.41, DC:monocyte-derived:anti-DC-SIGN_2h: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.4, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.4, DC:monocyte-derived:LPS: 0.4, DC:monocyte-derived:AEC-conditioned: 0.4, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.4, Macrophage:monocyte-derived:M-CSF/IFNg: 0.4, DC:monocyte-derived:Galectin-1: 0.4, DC:monocyte-derived: 0.4 |
| T188_AACTTCTCACAAACGG-1 | DC:monocyte-derived:anti-DC-SIGN_2h | 0.17 | 1509.15 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.39, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.39, DC:monocyte-derived:AEC-conditioned: 0.38, DC:monocyte-derived:Poly(IC): 0.38, DC:monocyte-derived:antiCD40/VAF347: 0.38, DC:monocyte-derived:LPS: 0.38, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.37, DC:monocyte-derived:Galectin-1: 0.37, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.37, Macrophage:monocyte-derived:M-CSF: 0.37 |
| T188_CTAACTTCATGGGAAC-1 | DC:monocyte-derived:anti-DC-SIGN_2h | 0.17 | 1433.18 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.41, DC:monocyte-derived:anti-DC-SIGN_2h: 0.41, Monocyte:leukotriene_D4: 0.4, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.4, Macrophage:monocyte-derived:M-CSF/Pam3Cys: 0.4, Macrophage:monocyte-derived:M-CSF/IFNg: 0.4, DC:monocyte-derived:LPS: 0.4, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.4, DC:monocyte-derived:antiCD40/VAF347: 0.39 |
| T188_TCACATTGTATCAGGG-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.16 | 1350.23 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.41, Macrophage:monocyte-derived:M-CSF: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.41, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg: 0.41, DC:monocyte-derived:LPS: 0.41, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.41, DC:monocyte-derived:AEC-conditioned: 0.41, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.41, DC:monocyte-derived:Poly(IC): 0.4 |
| T188_AGTTAGCGTCCGAAGA-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.18 | 1334.14 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.43, Macrophage:monocyte-derived:M-CSF/IFNg: 0.42, DC:monocyte-derived:anti-DC-SIGN_2h: 0.42, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.42, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.42, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.42, DC:monocyte-derived:AEC-conditioned: 0.42, Monocyte:leukotriene_D4: 0.41, DC:monocyte-derived:LPS: 0.41, DC:monocyte-derived: 0.41 |
| T188_TTCGATTCAATCAGCT-1 | DC:monocyte-derived:anti-DC-SIGN_2h | 0.18 | 1331.22 | Raw ScoresMacrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.44, Macrophage:monocyte-derived:M-CSF: 0.43, Macrophage:monocyte-derived:M-CSF/IFNg: 0.43, Monocyte:leukotriene_D4: 0.42, DC:monocyte-derived:anti-DC-SIGN_2h: 0.42, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.42, Monocyte:CD16-: 0.42, Macrophage:monocyte-derived:M-CSF/Pam3Cys: 0.42, Monocyte: 0.42, Monocyte:anti-FcgRIIB: 0.42 |
| T188_ATGCATGGTCGGCCTA-1 | Macrophage:monocyte-derived:IL-4/Dex/cntrl | 0.16 | 1215.33 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.4, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.4, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.39, Macrophage:monocyte-derived:M-CSF: 0.39, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.39, Monocyte:leukotriene_D4: 0.38, Monocyte:anti-FcgRIIB: 0.38, DC:monocyte-derived:antiCD40/VAF347: 0.38, DC:monocyte-derived: 0.38, DC:monocyte-derived:AEC-conditioned: 0.38 |
| T188_CTGAGGCGTAGCACAG-1 | Monocyte:CD16+ | 0.14 | 1195.24 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.4, DC:monocyte-derived:anti-DC-SIGN_2h: 0.4, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.4, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.4, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.4, Macrophage:monocyte-derived:M-CSF/IFNg: 0.39, DC:monocyte-derived:AEC-conditioned: 0.39, Monocyte:CD16-: 0.39, Monocyte:leukotriene_D4: 0.39, Monocyte:anti-FcgRIIB: 0.39 |
| T188_GAGTTGTAGACCATTC-1 | DC:monocyte-derived:anti-DC-SIGN_2h | 0.09 | 1176.93 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.33, DC:monocyte-derived:AEC-conditioned: 0.33, Macrophage:monocyte-derived: 0.33, Macrophage:monocyte-derived:M-CSF: 0.32, Macrophage:monocyte-derived:IFNa: 0.32, DC:monocyte-derived:immature: 0.32, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.32, Monocyte:leukotriene_D4: 0.32, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.32, DC:monocyte-derived:antiCD40/VAF347: 0.32 |
| T188_TGAGACTTCATACGAC-1 | DC:monocyte-derived:mature | 0.13 | 1143.14 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.4, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.4, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.39, Monocyte:leukotriene_D4: 0.39, Macrophage:monocyte-derived:M-CSF/IFNg: 0.39, DC:monocyte-derived:Poly(IC): 0.39, Monocyte: 0.38, Monocyte:anti-FcgRIIB: 0.38, Monocyte:CD16+: 0.38, DC:monocyte-derived:antiCD40/VAF347: 0.38 |
| T188_TGTCCACAGAACTGAT-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.15 | 1108.77 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.38, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.37, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.37, Macrophage:monocyte-derived:M-CSF: 0.37, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.37, Monocyte:leukotriene_D4: 0.37, Macrophage:monocyte-derived:M-CSF/Pam3Cys: 0.36, DC:monocyte-derived:AEC-conditioned: 0.36, DC:monocyte-derived: 0.36, Macrophage:monocyte-derived: 0.36 |
| T188_GATGCTAGTCTTCCGT-1 | Macrophage:monocyte-derived:IL-4/Dex/cntrl | 0.16 | 1103.73 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.42, DC:monocyte-derived:anti-DC-SIGN_2h: 0.41, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.41, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.41, Monocyte:leukotriene_D4: 0.4, Macrophage:monocyte-derived:M-CSF/IFNg: 0.4, DC:monocyte-derived:AEC-conditioned: 0.4, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.4, DC:monocyte-derived: 0.4, Macrophage:monocyte-derived:IL-4/cntrl: 0.39 |
| T188_CGGAATTCAAACCGGA-1 | DC:monocyte-derived:anti-DC-SIGN_2h | 0.18 | 1092.87 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.43, Macrophage:monocyte-derived:M-CSF: 0.42, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.42, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.42, Monocyte:leukotriene_D4: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg: 0.41, Monocyte: 0.41, DC:monocyte-derived: 0.41, DC:monocyte-derived:antiCD40/VAF347: 0.41 |
| T188_TCGGATAAGATTCGAA-1 | Macrophage:monocyte-derived:M-CSF/IFNg | 0.15 | 1053.30 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.41, Macrophage:monocyte-derived:M-CSF: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg: 0.4, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.4, Monocyte:leukotriene_D4: 0.4, DC:monocyte-derived:LPS: 0.4, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.4, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.4, DC:monocyte-derived:AEC-conditioned: 0.39, Monocyte: 0.39 |
| T188_CAATGACGTTCGAAGG-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.14 | 1013.31 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.4, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.39, Macrophage:monocyte-derived:M-CSF: 0.39, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.39, Monocyte:leukotriene_D4: 0.39, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.39, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.38, DC:monocyte-derived:antiCD40/VAF347: 0.38, Monocyte: 0.38, Monocyte:anti-FcgRIIB: 0.38 |
| T188_TCGTGCTAGAGGATCC-1 | DC:monocyte-derived:A._fumigatus_germ_tubes_6h | 0.18 | 1008.50 | Raw ScoresDC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.43, DC:monocyte-derived:anti-DC-SIGN_2h: 0.43, Macrophage:monocyte-derived:S._aureus: 0.42, Macrophage:monocyte-derived:M-CSF: 0.42, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.42, DC:monocyte-derived:Galectin-1: 0.42, DC:monocyte-derived:LPS: 0.42, DC:monocyte-derived: 0.42, Macrophage:Alveolar:B._anthacis_spores: 0.42, DC:monocyte-derived:antiCD40/VAF347: 0.42 |
| T188_CCGCAAGTCTTCGGAA-1 | DC:monocyte-derived:AEC-conditioned | 0.14 | 997.86 | Raw ScoresDC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.4, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.4, Macrophage:monocyte-derived:M-CSF/Pam3Cys: 0.39, DC:monocyte-derived:antiCD40/VAF347: 0.39, DC:monocyte-derived:anti-DC-SIGN_2h: 0.39, Macrophage:monocyte-derived:M-CSF: 0.39, Monocyte:anti-FcgRIIB: 0.38, Macrophage:monocyte-derived:M-CSF/IFNg: 0.38, DC:monocyte-derived:AEC-conditioned: 0.38, DC:monocyte-derived:LPS: 0.38 |
| T188_TCGACGGGTGATACCT-1 | Macrophage:monocyte-derived:M-CSF/IFNg | 0.16 | 950.19 | Raw ScoresMacrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.41, Macrophage:monocyte-derived:M-CSF: 0.4, Macrophage:monocyte-derived:M-CSF/IFNg: 0.4, DC:monocyte-derived:anti-DC-SIGN_2h: 0.4, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.4, Monocyte:leukotriene_D4: 0.39, DC:monocyte-derived:AEC-conditioned: 0.39, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.39, Macrophage:monocyte-derived:M-CSF/Pam3Cys: 0.39, DC:monocyte-derived:LPS: 0.39 |
| T188_TTACCATCAAGACGAC-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.15 | 932.46 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.39, Macrophage:monocyte-derived:M-CSF: 0.39, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.39, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.39, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.38, Monocyte:leukotriene_D4: 0.38, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.38, Macrophage:monocyte-derived:M-CSF/Pam3Cys: 0.38, Monocyte: 0.38, DC:monocyte-derived:AEC-conditioned: 0.38 |
| T188_TTGTTTGAGGCATCAG-1 | DC:monocyte-derived:anti-DC-SIGN_2h | 0.12 | 922.49 | Raw ScoresMonocyte:leukotriene_D4: 0.39, DC:monocyte-derived:anti-DC-SIGN_2h: 0.39, Monocyte: 0.38, NK_cell: 0.38, Monocyte:CD16+: 0.38, Pre-B_cell_CD34-: 0.38, Macrophage:monocyte-derived:M-CSF: 0.38, Monocyte:anti-FcgRIIB: 0.38, Monocyte:CD16-: 0.37, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.37 |
| T188_GTTGCTCCAGACCCGT-1 | Macrophage:monocyte-derived:IL-4/Dex/cntrl | 0.13 | 888.34 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.39, DC:monocyte-derived:anti-DC-SIGN_2h: 0.38, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.38, Monocyte:leukotriene_D4: 0.38, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.38, Macrophage:monocyte-derived:M-CSF/IFNg: 0.38, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.37, DC:monocyte-derived:AEC-conditioned: 0.37, Monocyte:anti-FcgRIIB: 0.37, Monocyte: 0.37 |
| T188_ACGGTCGCAAATGGTA-1 | Macrophage:monocyte-derived:M-CSF/IFNg | 0.18 | 883.93 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.41, DC:monocyte-derived:anti-DC-SIGN_2h: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.4, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.4, DC:monocyte-derived:LPS: 0.4, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.4, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.4, Monocyte:leukotriene_D4: 0.4, DC:monocyte-derived:AEC-conditioned: 0.39 |
| T188_TTTCAGTGTAATGCGG-1 | Macrophage:monocyte-derived:M-CSF | 0.21 | 872.59 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.43, Macrophage:monocyte-derived:M-CSF/IFNg: 0.42, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.41, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.41, Monocyte:leukotriene_D4: 0.41, DC:monocyte-derived:anti-DC-SIGN_2h: 0.41, DC:monocyte-derived:AEC-conditioned: 0.41, Monocyte:CD16-: 0.4, Monocyte:anti-FcgRIIB: 0.4, Macrophage:monocyte-derived: 0.4 |
| T188_GTTCCGTTCTCGTGAA-1 | Macrophage:monocyte-derived:M-CSF | 0.20 | 866.09 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.42, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.42, Macrophage:monocyte-derived:S._aureus: 0.41, Macrophage:monocyte-derived:IL-4/cntrl: 0.41, DC:monocyte-derived: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.41, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.41, Macrophage:monocyte-derived: 0.41, DC:monocyte-derived:antiCD40/VAF347: 0.41, Macrophage:monocyte-derived:IL-4/TGFb: 0.41 |
| T188_AAGGTAACATATCGGT-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.13 | 808.99 | Raw ScoresMacrophage:monocyte-derived:IL-4/Dex/cntrl: 0.37, Macrophage:monocyte-derived:M-CSF: 0.37, DC:monocyte-derived:anti-DC-SIGN_2h: 0.37, Monocyte:leukotriene_D4: 0.37, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.37, Macrophage:monocyte-derived:M-CSF/IFNg: 0.36, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.36, Monocyte:CD16-: 0.36, Monocyte: 0.36, Macrophage:monocyte-derived:M-CSF/Pam3Cys: 0.36 |
| T188_CCTCTCCTCCTGATAG-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.17 | 803.41 | Raw ScoresMacrophage:monocyte-derived:IL-4/Dex/TGFb: 0.42, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.42, DC:monocyte-derived:anti-DC-SIGN_2h: 0.42, Macrophage:monocyte-derived:M-CSF: 0.42, Monocyte:leukotriene_D4: 0.41, DC:monocyte-derived: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg: 0.4, DC:monocyte-derived:AEC-conditioned: 0.4, Macrophage:monocyte-derived:IL-4/cntrl: 0.4, Macrophage:monocyte-derived: 0.4 |
| T188_TGCACGGCAAAGCACG-1 | DC:monocyte-derived:anti-DC-SIGN_2h | 0.19 | 796.15 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.43, DC:monocyte-derived:anti-DC-SIGN_2h: 0.43, Macrophage:monocyte-derived:M-CSF/IFNg: 0.42, Monocyte:leukotriene_D4: 0.42, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.42, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.42, DC:monocyte-derived:AEC-conditioned: 0.42, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.41, Monocyte: 0.41, Monocyte:CD16-: 0.41 |
| T188_ATTCTTGCACACGGAA-1 | DC:monocyte-derived:anti-DC-SIGN_2h | 0.16 | 758.59 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.43, Monocyte:leukotriene_D4: 0.42, Macrophage:monocyte-derived:M-CSF/IFNg: 0.42, Monocyte:anti-FcgRIIB: 0.42, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.42, Monocyte: 0.42, DC:monocyte-derived:LPS: 0.41, Macrophage:monocyte-derived:M-CSF: 0.41, Monocyte:CD16+: 0.41, DC:monocyte-derived:Poly(IC): 0.41 |
| T188_GGCTTTCGTCAGTCTA-1 | DC:monocyte-derived:anti-DC-SIGN_2h | 0.11 | 711.65 | Raw ScoresMonocyte:leukotriene_D4: 0.37, DC:monocyte-derived:anti-DC-SIGN_2h: 0.37, Monocyte: 0.36, Monocyte:anti-FcgRIIB: 0.36, Macrophage:monocyte-derived:M-CSF: 0.36, Monocyte:CD16+: 0.36, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.36, Macrophage:monocyte-derived:M-CSF/IFNg: 0.35, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.35, Monocyte:CD16-: 0.35 |
| T230_GTGCTGGAGCCAGTAG-1 | Macrophage:monocyte-derived:M-CSF | 0.20 | 682.57 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.44, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.43, Macrophage:monocyte-derived:M-CSF: 0.43, DC:monocyte-derived: 0.43, DC:monocyte-derived:antiCD40/VAF347: 0.42, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.42, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.42, DC:monocyte-derived:Galectin-1: 0.42, DC:monocyte-derived:AEC-conditioned: 0.41, Macrophage:Alveolar:B._anthacis_spores: 0.41 |
| T188_CAGTGCGAGCCTGACC-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.16 | 661.04 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.41, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.4, DC:monocyte-derived:antiCD40/VAF347: 0.4, Macrophage:monocyte-derived:M-CSF: 0.4, DC:monocyte-derived: 0.4, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.4, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.4, Macrophage:monocyte-derived:S._aureus: 0.4, Macrophage:Alveolar:B._anthacis_spores: 0.4 |
| T188_TCACGCTTCCTACGGG-1 | DC:monocyte-derived:A._fumigatus_germ_tubes_6h | 0.19 | 629.31 | Raw ScoresDC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.44, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.43, DC:monocyte-derived:antiCD40/VAF347: 0.42, Macrophage:monocyte-derived:S._aureus: 0.42, Macrophage:monocyte-derived:M-CSF/Pam3Cys: 0.42, DC:monocyte-derived:Poly(IC): 0.42, DC:monocyte-derived:Schuler_treatment: 0.42, DC:monocyte-derived:LPS: 0.42, Macrophage:Alveolar:B._anthacis_spores: 0.41, DC:monocyte-derived:Galectin-1: 0.41 |
| T188_TCAAGCACATCGGTTA-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.14 | 602.59 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.42, DC:monocyte-derived:anti-DC-SIGN_2h: 0.42, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.41, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.41, Monocyte:leukotriene_D4: 0.41, Monocyte: 0.4, DC:monocyte-derived:AEC-conditioned: 0.4, DC:monocyte-derived: 0.4, Macrophage:monocyte-derived:M-CSF/IFNg: 0.4, Macrophage:monocyte-derived: 0.4 |
| T188_GAGTCTAAGGCACTCC-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.18 | 582.95 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg: 0.41, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.41, Monocyte:leukotriene_D4: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.41, DC:monocyte-derived:anti-DC-SIGN_2h: 0.41, DC:monocyte-derived:AEC-conditioned: 0.4, DC:monocyte-derived:antiCD40/VAF347: 0.4, Monocyte: 0.4, DC:monocyte-derived:LPS: 0.4 |
| T188_TTGCTGCAGGCGTCCT-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.16 | 539.88 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.42, Macrophage:monocyte-derived:M-CSF/IFNg: 0.41, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.41, Monocyte:leukotriene_D4: 0.41, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.41, DC:monocyte-derived:anti-DC-SIGN_2h: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.41, Monocyte: 0.4, Monocyte:CD16-: 0.4, Monocyte:CD16+: 0.4 |
| T230_AGTTCCCCACTACACA-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.21 | 538.59 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.45, DC:monocyte-derived:anti-DC-SIGN_2h: 0.45, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.45, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.44, DC:monocyte-derived: 0.44, Macrophage:monocyte-derived:M-CSF/IFNg: 0.43, DC:monocyte-derived:AEC-conditioned: 0.43, DC:monocyte-derived:LPS: 0.43, DC:monocyte-derived:Galectin-1: 0.43, Monocyte:leukotriene_D4: 0.43 |
| T188_ACATGCAAGCATCAAA-1 | DC:monocyte-derived:A._fumigatus_germ_tubes_6h | 0.16 | 536.58 | Raw ScoresMacrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.43, Monocyte:CD16-: 0.43, Monocyte:leukotriene_D4: 0.42, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.42, Monocyte: 0.42, Monocyte:anti-FcgRIIB: 0.42, Macrophage:monocyte-derived:M-CSF/IFNg: 0.42, DC:monocyte-derived:Schuler_treatment: 0.42, Macrophage:monocyte-derived:M-CSF: 0.42, Monocyte:CD14+: 0.42 |
| T188_GGAAGTGTCTGAACGT-1 | Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys | 0.22 | 531.61 | Raw ScoresMacrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.47, Monocyte:CD16-: 0.47, Macrophage:monocyte-derived:M-CSF/IFNg: 0.46, Monocyte:leukotriene_D4: 0.46, Macrophage:monocyte-derived:M-CSF/Pam3Cys: 0.46, Monocyte: 0.46, Macrophage:monocyte-derived:M-CSF: 0.46, Monocyte:CD14+: 0.46, Monocyte:anti-FcgRIIB: 0.46, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.45 |
| T188_GTTGCGGCATGATGCT-1 | Monocyte:CD16- | 0.17 | 521.98 | Raw ScoresMacrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.42, DC:monocyte-derived:anti-DC-SIGN_2h: 0.42, Monocyte:leukotriene_D4: 0.42, Macrophage:monocyte-derived:M-CSF/IFNg: 0.42, Monocyte:CD16-: 0.42, Monocyte: 0.42, Macrophage:monocyte-derived:M-CSF: 0.41, Monocyte:anti-FcgRIIB: 0.41, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.41, Monocyte:CD14+: 0.41 |
| T188_CTGTATTTCTCGTGGG-1 | Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys | 0.19 | 510.68 | Raw ScoresMacrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.45, Monocyte:CD16-: 0.44, Monocyte:leukotriene_D4: 0.44, Monocyte: 0.44, Monocyte:anti-FcgRIIB: 0.44, Macrophage:monocyte-derived:M-CSF/IFNg: 0.44, Macrophage:monocyte-derived:M-CSF/Pam3Cys: 0.44, Macrophage:monocyte-derived:M-CSF: 0.43, Monocyte:CD14+: 0.43, Monocyte:CD16+: 0.43 |
| T188_AGATCGTAGTCATTGC-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.14 | 506.80 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.38, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.37, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.37, Macrophage:monocyte-derived:M-CSF: 0.37, DC:monocyte-derived:AEC-conditioned: 0.36, DC:monocyte-derived: 0.36, Monocyte:leukotriene_D4: 0.36, DC:monocyte-derived:antiCD40/VAF347: 0.36, Macrophage:monocyte-derived:M-CSF/IFNg: 0.36, Macrophage:monocyte-derived: 0.36 |
| T214_AGACAAACAGTAACGG-1 | DC:monocyte-derived:Galectin-1 | 0.18 | 503.60 | Raw ScoresDC:monocyte-derived:Galectin-1: 0.41, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.41, DC:monocyte-derived:antiCD40/VAF347: 0.41, DC:monocyte-derived: 0.4, DC:monocyte-derived:anti-DC-SIGN_2h: 0.4, DC:monocyte-derived:LPS: 0.4, Macrophage:monocyte-derived: 0.4, DC:monocyte-derived:CD40L: 0.4, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.4, Macrophage:monocyte-derived:IL-4/cntrl: 0.4 |
| T188_CGTGATAAGCTCAGAG-1 | Macrophage:monocyte-derived:M-CSF/IFNg | 0.14 | 497.75 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.39, Macrophage:monocyte-derived:M-CSF: 0.38, DC:monocyte-derived:AEC-conditioned: 0.38, Macrophage:monocyte-derived:M-CSF/IFNg: 0.38, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.38, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.38, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.38, DC:monocyte-derived:LPS: 0.38, Monocyte:anti-FcgRIIB: 0.37, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.37 |
| T200_CCGGTGACAAATTGCC-1 | Macrophage:monocyte-derived:M-CSF/IFNg | 0.22 | 486.98 | Raw ScoresMacrophage:monocyte-derived:M-CSF/IFNg: 0.49, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.48, Macrophage:monocyte-derived:M-CSF: 0.48, Monocyte:leukotriene_D4: 0.48, Monocyte:anti-FcgRIIB: 0.47, Monocyte: 0.47, Monocyte:CD16-: 0.47, DC:monocyte-derived:LPS: 0.47, DC:monocyte-derived:AEC-conditioned: 0.47, Macrophage:monocyte-derived:M-CSF/Pam3Cys: 0.47 |
| T175_CTCCTCCTCCAGCTCT-1 | DC:monocyte-derived:anti-DC-SIGN_2h | 0.20 | 477.16 | Raw ScoresDC:monocyte-derived:anti-DC-SIGN_2h: 0.43, Macrophage:monocyte-derived:M-CSF: 0.43, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.42, Macrophage:monocyte-derived:M-CSF/IFNg: 0.42, Monocyte:leukotriene_D4: 0.42, DC:monocyte-derived: 0.42, DC:monocyte-derived:Galectin-1: 0.42, DC:monocyte-derived:LPS: 0.42, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.41, DC:monocyte-derived:antiCD40/VAF347: 0.41 |
| T188_AGTAGTCAGGGACAGG-1 | Macrophage:monocyte-derived:IL-4/Dex/TGFb | 0.14 | 473.08 | Raw ScoresMacrophage:monocyte-derived:M-CSF: 0.41, DC:monocyte-derived:anti-DC-SIGN_2h: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg: 0.4, Macrophage:monocyte-derived:IL-4/Dex/TGFb: 0.4, Macrophage:monocyte-derived:IL-4/Dex/cntrl: 0.4, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.4, Monocyte:leukotriene_D4: 0.4, DC:monocyte-derived:AEC-conditioned: 0.4, Monocyte:anti-FcgRIIB: 0.39, Monocyte:CD16-: 0.39 |
| T188_TGTGCGGCAATAGAGT-1 | Macrophage:monocyte-derived:M-CSF | 0.16 | 451.82 | Raw ScoresMonocyte:leukotriene_D4: 0.42, Macrophage:monocyte-derived:M-CSF: 0.42, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.41, Monocyte: 0.41, DC:monocyte-derived:anti-DC-SIGN_2h: 0.41, Macrophage:monocyte-derived:M-CSF/IFNg: 0.41, Monocyte:anti-FcgRIIB: 0.41, Monocyte:CD16-: 0.41, DC:monocyte-derived:AEC-conditioned: 0.41, DC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.4 |
| T40_CACCACTGTTCAACCA.1 | DC:monocyte-derived:anti-DC-SIGN_2h | 0.13 | 431.67 | Raw ScoresDC:monocyte-derived:A._fumigatus_germ_tubes_6h: 0.4, DC:monocyte-derived:anti-DC-SIGN_2h: 0.39, DC:monocyte-derived:antiCD40/VAF347: 0.38, DC:monocyte-derived: 0.38, Macrophage:monocyte-derived:M-CSF/IFNg/Pam3Cys: 0.38, DC:monocyte-derived:Galectin-1: 0.37, DC:monocyte-derived:Schuler_treatment: 0.37, DC:monocyte-derived:CD40L: 0.37, DC:monocyte-derived:AEC-conditioned: 0.37, DC:monocyte-derived:mature: 0.37 |
Below shows the significant enrichments of this GEP for literature curated gene lists
This data was procured from existing single cell RNA-seq maps of neuroblastoma or related relevant data.
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by the P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
M-MDSC
These marker genes were curated for MDSC subtypes as reviewed in Veglia et. al. (PMID 33526920):
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.28e-06
Mean rank of genes in gene set: 1989.4
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| IL1B | 0.0020540 | 129 | GTEx | DepMap | Descartes | 13.94 | 1643.16 |
| VEGFA | 0.0019267 | 146 | GTEx | DepMap | Descartes | 0.67 | 11.50 |
| CD84 | 0.0018015 | 156 | GTEx | DepMap | Descartes | 1.06 | 25.86 |
| TNF | 0.0010611 | 326 | GTEx | DepMap | Descartes | 1.54 | 176.43 |
| CD274 | 0.0010120 | 345 | GTEx | DepMap | Descartes | 0.07 | 3.55 |
| HIF1A | 0.0009753 | 356 | GTEx | DepMap | Descartes | 1.20 | 66.03 |
| STAT3 | 0.0007967 | 452 | GTEx | DepMap | Descartes | 1.48 | 59.73 |
| CD14 | 0.0007397 | 492 | GTEx | DepMap | Descartes | 8.21 | 960.97 |
| TNFRSF10B | 0.0006596 | 549 | GTEx | DepMap | Descartes | 0.27 | 13.50 |
| IL10 | 0.0005856 | 614 | GTEx | DepMap | Descartes | 0.73 | 65.92 |
| TGFB1 | 0.0003998 | 836 | GTEx | DepMap | Descartes | 1.12 | 97.91 |
| ARG1 | -0.0000225 | 4580 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| ARG2 | -0.0000251 | 4691 | GTEx | DepMap | Descartes | 0.15 | 13.43 |
| NOS2 | -0.0000276 | 4782 | GTEx | DepMap | Descartes | 0.00 | 0.13 |
| CD36 | -0.0002697 | 11387 | GTEx | DepMap | Descartes | 0.83 | 26.86 |
M1 Macrophage
These genes were collated from multiple sources:
Wilcoxon ranksum test P-value for gene set overrepresentation: 4.91e-06
Mean rank of genes in gene set: 1447.73
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CD80 | 0.0029989 | 72 | GTEx | DepMap | Descartes | 0.18 | 10.20 |
| ITGAX | 0.0026627 | 82 | GTEx | DepMap | Descartes | 0.61 | 25.87 |
| IL1B | 0.0020540 | 129 | GTEx | DepMap | Descartes | 13.94 | 1643.16 |
| IL18 | 0.0017774 | 165 | GTEx | DepMap | Descartes | 1.99 | 204.39 |
| CXCL16 | 0.0017690 | 167 | GTEx | DepMap | Descartes | 1.69 | 150.64 |
| TNF | 0.0010611 | 326 | GTEx | DepMap | Descartes | 1.54 | 176.43 |
| TLR4 | 0.0007460 | 486 | GTEx | DepMap | Descartes | 0.41 | 6.68 |
| CD14 | 0.0007397 | 492 | GTEx | DepMap | Descartes | 8.21 | 960.97 |
| CCL5 | 0.0000896 | 2060 | GTEx | DepMap | Descartes | 0.91 | 178.86 |
| CCL2 | -0.0000089 | 3971 | GTEx | DepMap | Descartes | 3.89 | 740.68 |
| IL33 | -0.0001028 | 7975 | GTEx | DepMap | Descartes | 0.03 | 1.80 |
Stress response (Kinker)
These marker genes were obtained in a pan cancer cell lines NMF analysis of scRNA-eq data in Kinker et al (PMID 33128048) - 10 metaprograms were recovered that manifest across cancers.:
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.68e-04
Mean rank of genes in gene set: 466
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ATF3 | 0.0083247 | 2 | GTEx | DepMap | Descartes | 6.63 | 688.64 |
| GADD45B | 0.0076151 | 5 | GTEx | DepMap | Descartes | 10.87 | 1247.25 |
| SQSTM1 | 0.0056690 | 14 | GTEx | DepMap | Descartes | 3.86 | 290.72 |
| DDIT3 | 0.0025313 | 91 | GTEx | DepMap | Descartes | 1.66 | 344.84 |
| GADD45A | 0.0000712 | 2218 | GTEx | DepMap | Descartes | 0.64 | 91.64 |
Below shows ranks on this GEP for literature curated gene lists for large gene sets
These include those reported as mesenchymal/adrenergic by Van Groningen et al.
High ranks indicate this gene is a driver of this GEP (note these results are not ordered).
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
VanGroningen Adrenergic Genes
Adrenergic marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 9098.35
Median rank of genes in gene set: 9988
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TUBB4B | 0.0022082 | 119 | GTEx | DepMap | Descartes | 2.55 | 259.23 |
| DAPK1 | 0.0011511 | 299 | GTEx | DepMap | Descartes | 0.45 | 14.66 |
| NFIL3 | 0.0007935 | 453 | GTEx | DepMap | Descartes | 0.49 | 48.69 |
| IRS2 | 0.0007183 | 507 | GTEx | DepMap | Descartes | 0.79 | 19.40 |
| LYN | 0.0004798 | 714 | GTEx | DepMap | Descartes | 0.98 | 34.65 |
| MYO5A | 0.0004567 | 749 | GTEx | DepMap | Descartes | 0.31 | 4.83 |
| CXCR4 | 0.0004156 | 821 | GTEx | DepMap | Descartes | 3.37 | 491.35 |
| RALGDS | 0.0003899 | 856 | GTEx | DepMap | Descartes | 0.78 | 23.79 |
| ATP6V1B2 | 0.0003117 | 995 | GTEx | DepMap | Descartes | 1.05 | 26.71 |
| CHML | 0.0002169 | 1262 | GTEx | DepMap | Descartes | 0.21 | 5.01 |
| NCOA7 | 0.0002043 | 1301 | GTEx | DepMap | Descartes | 1.06 | NA |
| PIK3R1 | 0.0001730 | 1457 | GTEx | DepMap | Descartes | 1.00 | 33.65 |
| FOXO3 | 0.0001322 | 1681 | GTEx | DepMap | Descartes | 0.96 | 20.70 |
| POLB | 0.0000813 | 2130 | GTEx | DepMap | Descartes | 0.17 | 23.94 |
| CELF2 | 0.0000802 | 2138 | GTEx | DepMap | Descartes | 0.78 | 21.53 |
| ARL6IP1 | 0.0000660 | 2285 | GTEx | DepMap | Descartes | 1.64 | 130.01 |
| RBBP8 | 0.0000582 | 2379 | GTEx | DepMap | Descartes | 0.14 | 8.37 |
| ST3GAL6 | 0.0000459 | 2534 | GTEx | DepMap | Descartes | 0.29 | 16.55 |
| CEP44 | 0.0000255 | 2900 | GTEx | DepMap | Descartes | 0.19 | 6.38 |
| CCSAP | 0.0000164 | 3119 | GTEx | DepMap | Descartes | 0.16 | NA |
| MYRIP | 0.0000157 | 3139 | GTEx | DepMap | Descartes | 0.01 | 0.27 |
| UCP2 | 0.0000138 | 3184 | GTEx | DepMap | Descartes | 2.33 | 202.75 |
| MMD | 0.0000118 | 3239 | GTEx | DepMap | Descartes | 0.56 | 36.96 |
| PEG3 | 0.0000070 | 3382 | GTEx | DepMap | Descartes | 0.13 | NA |
| CACNA2D2 | -0.0000018 | 3675 | GTEx | DepMap | Descartes | 0.03 | 0.89 |
| PRSS12 | -0.0000224 | 4574 | GTEx | DepMap | Descartes | 0.06 | 1.51 |
| RTN2 | -0.0000255 | 4704 | GTEx | DepMap | Descartes | 0.15 | 10.24 |
| FAM169A | -0.0000268 | 4750 | GTEx | DepMap | Descartes | 0.04 | 1.25 |
| ESRRG | -0.0000298 | 4876 | GTEx | DepMap | Descartes | 0.01 | 0.23 |
| TBC1D30 | -0.0000307 | 4911 | GTEx | DepMap | Descartes | 0.04 | 0.88 |
VanGroningen Mesenchymal Genes
Mesenchymal marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.42e-08
Mean rank of genes in gene set: 5343.67
Median rank of genes in gene set: 4947
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| SQSTM1 | 0.0056690 | 14 | GTEx | DepMap | Descartes | 3.86 | 290.72 |
| KLF4 | 0.0047289 | 24 | GTEx | DepMap | Descartes | 2.16 | 157.54 |
| KLF6 | 0.0046827 | 27 | GTEx | DepMap | Descartes | 9.10 | 471.40 |
| LMNA | 0.0039626 | 44 | GTEx | DepMap | Descartes | 3.74 | 242.94 |
| SGK1 | 0.0037918 | 51 | GTEx | DepMap | Descartes | 4.63 | 176.80 |
| FILIP1L | 0.0034746 | 55 | GTEx | DepMap | Descartes | 0.63 | 30.75 |
| PXDC1 | 0.0032368 | 66 | GTEx | DepMap | Descartes | 0.35 | 34.80 |
| HES1 | 0.0031291 | 68 | GTEx | DepMap | Descartes | 2.45 | 292.66 |
| SASH1 | 0.0029448 | 73 | GTEx | DepMap | Descartes | 0.43 | 11.49 |
| SDCBP | 0.0024594 | 102 | GTEx | DepMap | Descartes | 3.63 | 197.72 |
| CTSC | 0.0023946 | 108 | GTEx | DepMap | Descartes | 7.93 | 208.19 |
| APOE | 0.0022957 | 115 | GTEx | DepMap | Descartes | 104.15 | 15735.56 |
| RGL1 | 0.0020978 | 124 | GTEx | DepMap | Descartes | 0.41 | 16.10 |
| LITAF | 0.0019904 | 135 | GTEx | DepMap | Descartes | 3.44 | 262.84 |
| PDGFC | 0.0019630 | 140 | GTEx | DepMap | Descartes | 0.09 | 4.71 |
| BAG3 | 0.0019340 | 144 | GTEx | DepMap | Descartes | 0.41 | 33.24 |
| ITM2B | 0.0018226 | 155 | GTEx | DepMap | Descartes | 14.00 | 266.48 |
| KLF10 | 0.0017416 | 173 | GTEx | DepMap | Descartes | 0.97 | 55.80 |
| EGR1 | 0.0016851 | 183 | GTEx | DepMap | Descartes | 5.96 | 397.47 |
| INSIG1 | 0.0016015 | 198 | GTEx | DepMap | Descartes | 0.92 | 63.92 |
| GDF15 | 0.0015483 | 206 | GTEx | DepMap | Descartes | 0.23 | 44.78 |
| PLSCR1 | 0.0015060 | 215 | GTEx | DepMap | Descartes | 1.44 | 142.52 |
| TSPAN4 | 0.0014791 | 218 | GTEx | DepMap | Descartes | 1.36 | 87.62 |
| CXCL12 | 0.0013369 | 247 | GTEx | DepMap | Descartes | 0.74 | 39.55 |
| ARL4A | 0.0012695 | 269 | GTEx | DepMap | Descartes | 1.16 | 95.56 |
| MRC2 | 0.0012085 | 284 | GTEx | DepMap | Descartes | 0.23 | 9.05 |
| MYLIP | 0.0011772 | 293 | GTEx | DepMap | Descartes | 0.54 | 37.59 |
| EGR3 | 0.0010680 | 322 | GTEx | DepMap | Descartes | 0.48 | 23.09 |
| SKIL | 0.0010652 | 324 | GTEx | DepMap | Descartes | 0.92 | 30.47 |
| PAPSS2 | 0.0009503 | 366 | GTEx | DepMap | Descartes | 0.24 | 9.71 |
Descartes adrenocortical markers
Top 50 marker genes of adrenocortical cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/adrenocortical/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.90e-02
Mean rank of genes in gene set: 5498.97
Median rank of genes in gene set: 5605.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ERN1 | 0.0010830 | 313 | GTEx | DepMap | Descartes | 0.55 | 15.75 |
| SH3BP5 | 0.0009624 | 361 | GTEx | DepMap | Descartes | 0.64 | 38.32 |
| PAPSS2 | 0.0009503 | 366 | GTEx | DepMap | Descartes | 0.24 | 9.71 |
| APOC1 | 0.0008809 | 396 | GTEx | DepMap | Descartes | 39.63 | 10013.99 |
| LDLR | 0.0008727 | 403 | GTEx | DepMap | Descartes | 0.18 | 5.78 |
| SCARB1 | 0.0004347 | 781 | GTEx | DepMap | Descartes | 0.24 | 7.51 |
| HMGCS1 | 0.0003658 | 896 | GTEx | DepMap | Descartes | 0.24 | 9.09 |
| FDX1 | 0.0002502 | 1146 | GTEx | DepMap | Descartes | 0.67 | 44.06 |
| GRAMD1B | 0.0002091 | 1287 | GTEx | DepMap | Descartes | 0.08 | 2.34 |
| POR | 0.0001866 | 1383 | GTEx | DepMap | Descartes | 0.47 | 38.29 |
| DHCR7 | 0.0000659 | 2289 | GTEx | DepMap | Descartes | 0.08 | 5.86 |
| NPC1 | 0.0000157 | 3140 | GTEx | DepMap | Descartes | 0.17 | 7.13 |
| FREM2 | 0.0000116 | 3247 | GTEx | DepMap | Descartes | 0.00 | 0.01 |
| STAR | 0.0000076 | 3363 | GTEx | DepMap | Descartes | 0.03 | 2.77 |
| PEG3 | 0.0000070 | 3382 | GTEx | DepMap | Descartes | 0.13 | NA |
| SGCZ | 0.0000007 | 3592 | GTEx | DepMap | Descartes | 0.00 | 0.02 |
| SH3PXD2B | -0.0000049 | 3806 | GTEx | DepMap | Descartes | 0.10 | 2.36 |
| SLC16A9 | -0.0000377 | 5217 | GTEx | DepMap | Descartes | 0.04 | 1.49 |
| INHA | -0.0000558 | 5994 | GTEx | DepMap | Descartes | 0.01 | 1.70 |
| BAIAP2L1 | -0.0000564 | 6021 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PDE10A | -0.0000725 | 6700 | GTEx | DepMap | Descartes | 0.05 | 0.99 |
| DHCR24 | -0.0000766 | 6865 | GTEx | DepMap | Descartes | 0.06 | 1.32 |
| FRMD5 | -0.0000819 | 7098 | GTEx | DepMap | Descartes | 0.01 | 0.30 |
| SLC1A2 | -0.0000930 | 7602 | GTEx | DepMap | Descartes | 0.05 | 0.46 |
| FDXR | -0.0000988 | 7808 | GTEx | DepMap | Descartes | 0.09 | 6.45 |
| HMGCR | -0.0001065 | 8110 | GTEx | DepMap | Descartes | 0.11 | 4.89 |
| TM7SF2 | -0.0001095 | 8222 | GTEx | DepMap | Descartes | 0.20 | 14.49 |
| IGF1R | -0.0001182 | 8516 | GTEx | DepMap | Descartes | 0.16 | 2.17 |
| SCAP | -0.0001264 | 8800 | GTEx | DepMap | Descartes | 0.16 | 6.10 |
| JAKMIP2 | -0.0001692 | 9957 | GTEx | DepMap | Descartes | 0.10 | 1.70 |
Descartes chromaffin markers
Top 50 marker genes of chromaffin cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/chromaffin/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 9317.9
Median rank of genes in gene set: 10335
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| EYA4 | 0.0000196 | 3040 | GTEx | DepMap | Descartes | 0.05 | 1.38 |
| SLC44A5 | -0.0000112 | 4072 | GTEx | DepMap | Descartes | 0.01 | 0.51 |
| RYR2 | -0.0000194 | 4425 | GTEx | DepMap | Descartes | 0.04 | 0.32 |
| PTCHD1 | -0.0000301 | 4888 | GTEx | DepMap | Descartes | 0.04 | 0.42 |
| PLXNA4 | -0.0000350 | 5103 | GTEx | DepMap | Descartes | 0.08 | 0.83 |
| EPHA6 | -0.0000490 | 5699 | GTEx | DepMap | Descartes | 0.01 | 0.30 |
| HS3ST5 | -0.0000538 | 5908 | GTEx | DepMap | Descartes | 0.03 | 1.27 |
| ELAVL2 | -0.0000594 | 6164 | GTEx | DepMap | Descartes | 0.24 | 7.06 |
| RPH3A | -0.0000628 | 6289 | GTEx | DepMap | Descartes | 0.02 | 0.80 |
| ANKFN1 | -0.0000714 | 6642 | GTEx | DepMap | Descartes | 0.01 | 0.34 |
| FAT3 | -0.0000751 | 6807 | GTEx | DepMap | Descartes | 0.04 | 0.41 |
| RBFOX1 | -0.0000817 | 7087 | GTEx | DepMap | Descartes | 0.10 | 2.73 |
| ALK | -0.0001015 | 7928 | GTEx | DepMap | Descartes | 0.03 | 0.80 |
| SYNPO2 | -0.0001068 | 8125 | GTEx | DepMap | Descartes | 0.22 | 2.03 |
| KCNB2 | -0.0001214 | 8636 | GTEx | DepMap | Descartes | 0.01 | 0.54 |
| CNKSR2 | -0.0001291 | 8878 | GTEx | DepMap | Descartes | 0.04 | 0.52 |
| MAB21L2 | -0.0001306 | 8924 | GTEx | DepMap | Descartes | 0.25 | 10.84 |
| EYA1 | -0.0001429 | 9287 | GTEx | DepMap | Descartes | 0.05 | 1.60 |
| BASP1 | -0.0001453 | 9346 | GTEx | DepMap | Descartes | 4.25 | 373.05 |
| GAL | -0.0001606 | 9759 | GTEx | DepMap | Descartes | 0.33 | 55.35 |
| MARCH11 | -0.0001904 | 10335 | GTEx | DepMap | Descartes | 0.56 | NA |
| RGMB | -0.0001912 | 10355 | GTEx | DepMap | Descartes | 0.21 | 6.83 |
| SLC6A2 | -0.0002071 | 10634 | GTEx | DepMap | Descartes | 0.10 | 3.99 |
| GAP43 | -0.0002142 | 10743 | GTEx | DepMap | Descartes | 1.17 | 80.72 |
| REEP1 | -0.0002188 | 10801 | GTEx | DepMap | Descartes | 0.11 | 3.33 |
| MAB21L1 | -0.0002238 | 10865 | GTEx | DepMap | Descartes | 0.26 | 11.89 |
| NPY | -0.0002656 | 11342 | GTEx | DepMap | Descartes | 4.99 | 834.89 |
| TMEM132C | -0.0003026 | 11634 | GTEx | DepMap | Descartes | 0.05 | 1.68 |
| TMEFF2 | -0.0003033 | 11637 | GTEx | DepMap | Descartes | 0.14 | 6.34 |
| IL7 | -0.0003301 | 11800 | GTEx | DepMap | Descartes | 0.12 | 9.87 |
Descartes Vascular_endothelial markers
Top 50 marker genes of Vascular_endothelial cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/vascular_endothelial/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.81e-01
Mean rank of genes in gene set: 5935.45
Median rank of genes in gene set: 6433
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CRHBP | 0.0010991 | 310 | GTEx | DepMap | Descartes | 0.12 | 14.08 |
| TMEM88 | 0.0006794 | 536 | GTEx | DepMap | Descartes | 0.18 | 46.02 |
| SHE | 0.0006473 | 560 | GTEx | DepMap | Descartes | 0.03 | 0.89 |
| ID1 | 0.0001722 | 1465 | GTEx | DepMap | Descartes | 0.45 | 76.45 |
| F8 | 0.0001262 | 1724 | GTEx | DepMap | Descartes | 0.02 | 0.34 |
| MYRIP | 0.0000157 | 3139 | GTEx | DepMap | Descartes | 0.01 | 0.27 |
| PTPRB | 0.0000152 | 3152 | GTEx | DepMap | Descartes | 0.09 | 0.74 |
| EHD3 | -0.0000096 | 4003 | GTEx | DepMap | Descartes | 0.03 | 0.76 |
| CYP26B1 | -0.0000098 | 4012 | GTEx | DepMap | Descartes | 0.03 | 0.83 |
| HYAL2 | -0.0000104 | 4032 | GTEx | DepMap | Descartes | 0.39 | 18.11 |
| PLVAP | -0.0000110 | 4062 | GTEx | DepMap | Descartes | 0.31 | 12.58 |
| GALNT15 | -0.0000140 | 4201 | GTEx | DepMap | Descartes | 0.01 | NA |
| SLCO2A1 | -0.0000298 | 4877 | GTEx | DepMap | Descartes | 0.02 | 0.42 |
| ESM1 | -0.0000353 | 5113 | GTEx | DepMap | Descartes | 0.10 | 3.07 |
| IRX3 | -0.0000398 | 5302 | GTEx | DepMap | Descartes | 0.02 | 1.36 |
| CDH13 | -0.0000417 | 5382 | GTEx | DepMap | Descartes | 0.05 | 0.62 |
| NOTCH4 | -0.0000487 | 5685 | GTEx | DepMap | Descartes | 0.13 | 2.58 |
| CEACAM1 | -0.0000539 | 5915 | GTEx | DepMap | Descartes | 0.01 | 0.61 |
| EFNB2 | -0.0000654 | 6412 | GTEx | DepMap | Descartes | 0.20 | 4.69 |
| ARHGAP29 | -0.0000663 | 6454 | GTEx | DepMap | Descartes | 0.24 | 3.98 |
| CDH5 | -0.0000669 | 6481 | GTEx | DepMap | Descartes | 0.10 | 2.73 |
| KDR | -0.0000724 | 6695 | GTEx | DepMap | Descartes | 0.11 | 1.33 |
| CALCRL | -0.0000748 | 6786 | GTEx | DepMap | Descartes | 0.07 | 1.28 |
| FLT4 | -0.0000836 | 7174 | GTEx | DepMap | Descartes | 0.02 | 0.57 |
| PODXL | -0.0000878 | 7357 | GTEx | DepMap | Descartes | 0.17 | 2.29 |
| CHRM3 | -0.0000924 | 7583 | GTEx | DepMap | Descartes | 0.10 | 1.72 |
| TIE1 | -0.0000968 | 7735 | GTEx | DepMap | Descartes | 0.05 | 1.70 |
| RASIP1 | -0.0001099 | 8238 | GTEx | DepMap | Descartes | 0.03 | 0.81 |
| MMRN2 | -0.0001127 | 8330 | GTEx | DepMap | Descartes | 0.04 | 0.95 |
| TEK | -0.0001145 | 8398 | GTEx | DepMap | Descartes | 0.01 | 0.20 |
Descartes stromal markers
Top 50 marker genes of stromal cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/stromal/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.74e-01
Mean rank of genes in gene set: 7334.32
Median rank of genes in gene set: 7638.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ABCA6 | 0.0008311 | 426 | GTEx | DepMap | Descartes | 0.11 | 2.65 |
| DKK2 | 0.0006060 | 592 | GTEx | DepMap | Descartes | 0.03 | 1.51 |
| COL27A1 | 0.0001303 | 1692 | GTEx | DepMap | Descartes | 0.02 | 0.38 |
| EDNRA | 0.0000509 | 2462 | GTEx | DepMap | Descartes | 0.06 | 2.73 |
| LAMC3 | -0.0000120 | 4113 | GTEx | DepMap | Descartes | 0.00 | 0.12 |
| ADAMTSL3 | -0.0000139 | 4195 | GTEx | DepMap | Descartes | 0.01 | 0.26 |
| FREM1 | -0.0000218 | 4543 | GTEx | DepMap | Descartes | 0.01 | 0.19 |
| PDGFRA | -0.0000323 | 4975 | GTEx | DepMap | Descartes | 0.11 | 2.82 |
| ITGA11 | -0.0000376 | 5214 | GTEx | DepMap | Descartes | 0.01 | 0.21 |
| POSTN | -0.0000490 | 5697 | GTEx | DepMap | Descartes | 0.45 | 14.92 |
| ABCC9 | -0.0000501 | 5744 | GTEx | DepMap | Descartes | 0.02 | 0.43 |
| GLI2 | -0.0000527 | 5858 | GTEx | DepMap | Descartes | 0.00 | 0.12 |
| ELN | -0.0000626 | 6283 | GTEx | DepMap | Descartes | 0.06 | 2.04 |
| BICC1 | -0.0000655 | 6414 | GTEx | DepMap | Descartes | 0.06 | 1.92 |
| CLDN11 | -0.0000706 | 6613 | GTEx | DepMap | Descartes | 0.02 | 1.30 |
| CDH11 | -0.0000732 | 6725 | GTEx | DepMap | Descartes | 0.09 | 2.17 |
| COL12A1 | -0.0000778 | 6919 | GTEx | DepMap | Descartes | 0.10 | 1.03 |
| PCDH18 | -0.0000789 | 6964 | GTEx | DepMap | Descartes | 0.02 | 0.56 |
| GAS2 | -0.0000799 | 7004 | GTEx | DepMap | Descartes | 0.00 | 0.38 |
| PRICKLE1 | -0.0000819 | 7093 | GTEx | DepMap | Descartes | 0.07 | 2.10 |
| PRRX1 | -0.0000845 | 7203 | GTEx | DepMap | Descartes | 0.08 | 3.20 |
| MGP | -0.0000929 | 7599 | GTEx | DepMap | Descartes | 1.76 | 190.69 |
| COL6A3 | -0.0000950 | 7678 | GTEx | DepMap | Descartes | 0.29 | 4.19 |
| LRRC17 | -0.0000998 | 7861 | GTEx | DepMap | Descartes | 0.02 | 2.27 |
| RSPO3 | -0.0001008 | 7893 | GTEx | DepMap | Descartes | 0.02 | NA |
| LOX | -0.0001045 | 8031 | GTEx | DepMap | Descartes | 0.08 | 1.91 |
| C7 | -0.0001139 | 8373 | GTEx | DepMap | Descartes | 0.10 | 3.37 |
| HHIP | -0.0001179 | 8505 | GTEx | DepMap | Descartes | 0.03 | 0.46 |
| ISLR | -0.0001328 | 9005 | GTEx | DepMap | Descartes | 0.10 | 8.00 |
| COL1A1 | -0.0001331 | 9012 | GTEx | DepMap | Descartes | 2.35 | 57.96 |
Descartes sympathoblasts markers
Top 50 marker genes of sympathoblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/sympathoblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 7.34e-01
Mean rank of genes in gene set: 6641.89
Median rank of genes in gene set: 6835
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ARC | 0.0004278 | 801 | GTEx | DepMap | Descartes | 0.82 | 48.47 |
| ST18 | 0.0001188 | 1786 | GTEx | DepMap | Descartes | 0.03 | 0.80 |
| CCSER1 | 0.0001084 | 1883 | GTEx | DepMap | Descartes | 0.04 | NA |
| CNTN3 | 0.0000648 | 2300 | GTEx | DepMap | Descartes | 0.01 | 0.49 |
| SLC24A2 | 0.0000322 | 2760 | GTEx | DepMap | Descartes | 0.01 | 0.24 |
| PENK | 0.0000236 | 2937 | GTEx | DepMap | Descartes | 0.05 | 5.61 |
| LAMA3 | 0.0000154 | 3146 | GTEx | DepMap | Descartes | 0.01 | 0.21 |
| GRM7 | 0.0000038 | 3477 | GTEx | DepMap | Descartes | 0.00 | 0.03 |
| MGAT4C | 0.0000008 | 3587 | GTEx | DepMap | Descartes | 0.04 | 0.20 |
| CDH12 | -0.0000103 | 4031 | GTEx | DepMap | Descartes | 0.00 | 0.21 |
| SORCS3 | -0.0000109 | 4059 | GTEx | DepMap | Descartes | 0.00 | 0.12 |
| ROBO1 | -0.0000279 | 4797 | GTEx | DepMap | Descartes | 0.12 | 2.42 |
| TBX20 | -0.0000343 | 5068 | GTEx | DepMap | Descartes | 0.00 | 0.21 |
| EML6 | -0.0000347 | 5086 | GTEx | DepMap | Descartes | 0.03 | 0.63 |
| FGF14 | -0.0000410 | 5354 | GTEx | DepMap | Descartes | 0.05 | 0.69 |
| CDH18 | -0.0000468 | 5597 | GTEx | DepMap | Descartes | 0.01 | 0.43 |
| DGKK | -0.0000505 | 5760 | GTEx | DepMap | Descartes | 0.02 | 0.42 |
| KSR2 | -0.0000569 | 6046 | GTEx | DepMap | Descartes | 0.05 | 0.37 |
| KCTD16 | -0.0000721 | 6673 | GTEx | DepMap | Descartes | 0.10 | 1.07 |
| GCH1 | -0.0000798 | 6997 | GTEx | DepMap | Descartes | 0.39 | 26.92 |
| NTNG1 | -0.0000822 | 7113 | GTEx | DepMap | Descartes | 0.06 | 1.71 |
| GRID2 | -0.0000857 | 7264 | GTEx | DepMap | Descartes | 0.03 | 0.71 |
| PCSK2 | -0.0000883 | 7383 | GTEx | DepMap | Descartes | 0.03 | 1.28 |
| TENM1 | -0.0000886 | 7402 | GTEx | DepMap | Descartes | 0.01 | NA |
| TIAM1 | -0.0001062 | 8100 | GTEx | DepMap | Descartes | 0.14 | 3.82 |
| SLC35F3 | -0.0001120 | 8306 | GTEx | DepMap | Descartes | 0.01 | 1.19 |
| GALNTL6 | -0.0001121 | 8310 | GTEx | DepMap | Descartes | 0.01 | 0.40 |
| FAM155A | -0.0001297 | 8900 | GTEx | DepMap | Descartes | 0.09 | 1.28 |
| UNC80 | -0.0001411 | 9235 | GTEx | DepMap | Descartes | 0.07 | 0.79 |
| AGBL4 | -0.0001435 | 9297 | GTEx | DepMap | Descartes | 0.01 | 0.70 |
Descartes erythroblasts markers
Top 50 marker genes of erythroblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/erythroblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.53e-01
Mean rank of genes in gene set: 5588.14
Median rank of genes in gene set: 6165
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| BLVRB | 0.0017557 | 169 | GTEx | DepMap | Descartes | 3.10 | 410.29 |
| GCLC | 0.0005126 | 673 | GTEx | DepMap | Descartes | 0.23 | 11.69 |
| ABCB10 | 0.0003417 | 942 | GTEx | DepMap | Descartes | 0.06 | 3.27 |
| CAT | 0.0002406 | 1173 | GTEx | DepMap | Descartes | 0.61 | 51.58 |
| DENND4A | 0.0002062 | 1295 | GTEx | DepMap | Descartes | 0.26 | 6.41 |
| SELENBP1 | 0.0001959 | 1332 | GTEx | DepMap | Descartes | 0.04 | 2.17 |
| MARCH3 | 0.0001821 | 1404 | GTEx | DepMap | Descartes | 0.12 | NA |
| RAPGEF2 | 0.0001106 | 1861 | GTEx | DepMap | Descartes | 0.31 | 7.18 |
| SLC25A21 | -0.0000007 | 3642 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SLC4A1 | -0.0000063 | 3858 | GTEx | DepMap | Descartes | 0.00 | 0.33 |
| SPECC1 | -0.0000084 | 3952 | GTEx | DepMap | Descartes | 0.15 | 3.90 |
| RHD | -0.0000229 | 4602 | GTEx | DepMap | Descartes | 0.00 | 0.14 |
| ALAS2 | -0.0000297 | 4873 | GTEx | DepMap | Descartes | 0.00 | 0.08 |
| SPTB | -0.0000470 | 5614 | GTEx | DepMap | Descartes | 0.02 | 0.21 |
| SOX6 | -0.0000594 | 6165 | GTEx | DepMap | Descartes | 0.02 | 0.41 |
| TFR2 | -0.0000691 | 6549 | GTEx | DepMap | Descartes | 0.03 | 1.77 |
| CPOX | -0.0000702 | 6597 | GTEx | DepMap | Descartes | 0.06 | 3.99 |
| MICAL2 | -0.0000728 | 6713 | GTEx | DepMap | Descartes | 0.07 | 2.37 |
| TMCC2 | -0.0000786 | 6951 | GTEx | DepMap | Descartes | 0.03 | 0.74 |
| RGS6 | -0.0000806 | 7034 | GTEx | DepMap | Descartes | 0.00 | 0.03 |
| ANK1 | -0.0000860 | 7282 | GTEx | DepMap | Descartes | 0.03 | 0.53 |
| XPO7 | -0.0000980 | 7773 | GTEx | DepMap | Descartes | 0.13 | 4.25 |
| TRAK2 | -0.0001295 | 8895 | GTEx | DepMap | Descartes | 0.11 | 2.72 |
| SNCA | -0.0001370 | 9112 | GTEx | DepMap | Descartes | 0.27 | 15.39 |
| SLC25A37 | -0.0001605 | 9755 | GTEx | DepMap | Descartes | 0.34 | 15.59 |
| GYPC | -0.0001825 | 10194 | GTEx | DepMap | Descartes | 1.18 | 147.95 |
| EPB41 | -0.0002158 | 10761 | GTEx | DepMap | Descartes | 0.25 | 10.02 |
| FECH | -0.0002253 | 10891 | GTEx | DepMap | Descartes | 0.11 | 2.47 |
| TSPAN5 | -0.0003723 | 11994 | GTEx | DepMap | Descartes | 0.20 | 8.67 |
| NA | NA | NA | NA | NA | NA | NA | NA |
Descartes myeloid markers
Top 50 marker genes of myeloid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/myeloid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.89e-16
Mean rank of genes in gene set: 1495.42
Median rank of genes in gene set: 550
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| FGL2 | 0.0051679 | 19 | GTEx | DepMap | Descartes | 3.13 | 134.57 |
| MS4A4A | 0.0045853 | 30 | GTEx | DepMap | Descartes | 2.94 | 310.97 |
| SLC1A3 | 0.0039586 | 45 | GTEx | DepMap | Descartes | 0.50 | 27.11 |
| LGMN | 0.0037568 | 53 | GTEx | DepMap | Descartes | 5.78 | 516.29 |
| MERTK | 0.0030976 | 69 | GTEx | DepMap | Descartes | 0.51 | 26.45 |
| SLCO2B1 | 0.0024898 | 95 | GTEx | DepMap | Descartes | 0.99 | 33.71 |
| CTSC | 0.0023946 | 108 | GTEx | DepMap | Descartes | 7.93 | 208.19 |
| RGL1 | 0.0020978 | 124 | GTEx | DepMap | Descartes | 0.41 | 16.10 |
| IFNGR1 | 0.0019495 | 142 | GTEx | DepMap | Descartes | 1.44 | 113.44 |
| ABCA1 | 0.0017949 | 158 | GTEx | DepMap | Descartes | 1.15 | 22.86 |
| CSF1R | 0.0014660 | 219 | GTEx | DepMap | Descartes | 1.38 | 70.98 |
| CD163L1 | 0.0014039 | 234 | GTEx | DepMap | Descartes | 0.40 | 16.08 |
| ADAP2 | 0.0013704 | 238 | GTEx | DepMap | Descartes | 0.99 | 74.02 |
| WWP1 | 0.0013444 | 244 | GTEx | DepMap | Descartes | 0.42 | 15.61 |
| PTPRE | 0.0012883 | 259 | GTEx | DepMap | Descartes | 1.00 | 34.78 |
| CD163 | 0.0011716 | 294 | GTEx | DepMap | Descartes | 1.47 | 57.46 |
| AXL | 0.0010197 | 343 | GTEx | DepMap | Descartes | 0.92 | 41.05 |
| SLC9A9 | 0.0009836 | 354 | GTEx | DepMap | Descartes | 0.26 | 15.09 |
| CD14 | 0.0007397 | 492 | GTEx | DepMap | Descartes | 8.21 | 960.97 |
| ITPR2 | 0.0005900 | 608 | GTEx | DepMap | Descartes | 0.54 | 8.95 |
| CTSS | 0.0005885 | 610 | GTEx | DepMap | Descartes | 6.44 | 276.55 |
| TGFBI | 0.0005050 | 681 | GTEx | DepMap | Descartes | 1.47 | 63.14 |
| SFMBT2 | 0.0004860 | 708 | GTEx | DepMap | Descartes | 0.21 | 5.33 |
| CTSB | 0.0004612 | 741 | GTEx | DepMap | Descartes | 13.03 | 693.84 |
| CYBB | 0.0004596 | 744 | GTEx | DepMap | Descartes | 1.49 | 64.36 |
| FMN1 | 0.0004377 | 776 | GTEx | DepMap | Descartes | 0.15 | 2.40 |
| CTSD | 0.0003779 | 876 | GTEx | DepMap | Descartes | 17.76 | 1715.65 |
| SPP1 | 0.0003173 | 981 | GTEx | DepMap | Descartes | 53.00 | 8630.67 |
| CPVL | 0.0003159 | 985 | GTEx | DepMap | Descartes | 2.61 | 199.96 |
| ATP8B4 | 0.0002955 | 1035 | GTEx | DepMap | Descartes | 0.13 | 5.42 |
Descartes Schwann markers
Top 50 marker genes of Schwann cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/schwann/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 5.75e-01
Mean rank of genes in gene set: 6378.77
Median rank of genes in gene set: 6415.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| PAG1 | 0.0004910 | 699 | GTEx | DepMap | Descartes | 0.46 | 9.41 |
| STARD13 | 0.0004666 | 731 | GTEx | DepMap | Descartes | 0.13 | 4.57 |
| KCTD12 | 0.0001910 | 1358 | GTEx | DepMap | Descartes | 0.82 | 26.61 |
| GAS7 | 0.0001655 | 1506 | GTEx | DepMap | Descartes | 0.24 | 6.13 |
| DST | 0.0000641 | 2309 | GTEx | DepMap | Descartes | 1.14 | 8.16 |
| MDGA2 | 0.0000404 | 2609 | GTEx | DepMap | Descartes | 0.00 | 0.22 |
| IL1RAPL2 | 0.0000354 | 2705 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| TRPM3 | 0.0000334 | 2740 | GTEx | DepMap | Descartes | 0.01 | 0.07 |
| ADAMTS5 | 0.0000123 | 3224 | GTEx | DepMap | Descartes | 0.04 | 0.36 |
| OLFML2A | 0.0000099 | 3296 | GTEx | DepMap | Descartes | 0.07 | 1.71 |
| IL1RAPL1 | 0.0000030 | 3507 | GTEx | DepMap | Descartes | 0.00 | 0.35 |
| HMGA2 | 0.0000017 | 3560 | GTEx | DepMap | Descartes | 0.00 | 0.13 |
| PPP2R2B | -0.0000141 | 4208 | GTEx | DepMap | Descartes | 0.11 | 1.28 |
| COL25A1 | -0.0000180 | 4366 | GTEx | DepMap | Descartes | 0.01 | 0.10 |
| PLP1 | -0.0000287 | 4840 | GTEx | DepMap | Descartes | 0.22 | 15.60 |
| PLCE1 | -0.0000320 | 4965 | GTEx | DepMap | Descartes | 0.04 | 0.52 |
| SLC35F1 | -0.0000339 | 5046 | GTEx | DepMap | Descartes | 0.02 | 0.77 |
| NRXN3 | -0.0000359 | 5137 | GTEx | DepMap | Descartes | 0.02 | 0.35 |
| XKR4 | -0.0000411 | 5356 | GTEx | DepMap | Descartes | 0.02 | 0.20 |
| PTPRZ1 | -0.0000471 | 5620 | GTEx | DepMap | Descartes | 0.00 | 0.05 |
| GRIK3 | -0.0000482 | 5669 | GTEx | DepMap | Descartes | 0.01 | 0.18 |
| ERBB4 | -0.0000614 | 6237 | GTEx | DepMap | Descartes | 0.01 | 0.13 |
| EGFLAM | -0.0000702 | 6594 | GTEx | DepMap | Descartes | 0.06 | 1.65 |
| SORCS1 | -0.0000732 | 6728 | GTEx | DepMap | Descartes | 0.05 | 1.00 |
| ERBB3 | -0.0000821 | 7105 | GTEx | DepMap | Descartes | 0.01 | 0.54 |
| EDNRB | -0.0000821 | 7106 | GTEx | DepMap | Descartes | 0.12 | 4.95 |
| LRRTM4 | -0.0000851 | 7233 | GTEx | DepMap | Descartes | 0.01 | 0.44 |
| COL18A1 | -0.0000874 | 7341 | GTEx | DepMap | Descartes | 0.41 | 9.51 |
| SOX5 | -0.0000879 | 7360 | GTEx | DepMap | Descartes | 0.08 | 1.63 |
| NRXN1 | -0.0000882 | 7377 | GTEx | DepMap | Descartes | 0.34 | 5.15 |
Descartes Megakaryocytes markers
Top 50 marker genes of Megakaryocytes cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/megakaryocytes/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 3.91e-02
Mean rank of genes in gene set: 5325.27
Median rank of genes in gene set: 4450
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| PLEK | 0.0020105 | 134 | GTEx | DepMap | Descartes | 2.56 | 190.78 |
| CD84 | 0.0018015 | 156 | GTEx | DepMap | Descartes | 1.06 | 25.86 |
| FERMT3 | 0.0010398 | 333 | GTEx | DepMap | Descartes | 0.76 | 57.34 |
| MCTP1 | 0.0006019 | 599 | GTEx | DepMap | Descartes | 0.14 | 4.37 |
| LIMS1 | 0.0005512 | 638 | GTEx | DepMap | Descartes | 2.01 | 86.89 |
| TGFB1 | 0.0003998 | 836 | GTEx | DepMap | Descartes | 1.12 | 97.91 |
| PSTPIP2 | 0.0003179 | 979 | GTEx | DepMap | Descartes | 0.16 | 10.65 |
| TLN1 | 0.0003018 | 1019 | GTEx | DepMap | Descartes | 1.03 | 22.06 |
| UBASH3B | 0.0003007 | 1021 | GTEx | DepMap | Descartes | 0.12 | 4.18 |
| RAP1B | 0.0001882 | 1372 | GTEx | DepMap | Descartes | 1.30 | 19.34 |
| TPM4 | 0.0001632 | 1515 | GTEx | DepMap | Descartes | 2.38 | 97.25 |
| STON2 | 0.0000817 | 2124 | GTEx | DepMap | Descartes | 0.08 | 2.83 |
| ARHGAP6 | 0.0000604 | 2348 | GTEx | DepMap | Descartes | 0.03 | 0.72 |
| TUBB1 | 0.0000361 | 2688 | GTEx | DepMap | Descartes | 0.00 | 0.32 |
| THBS1 | 0.0000335 | 2739 | GTEx | DepMap | Descartes | 0.53 | 10.77 |
| ITGB3 | 0.0000301 | 2805 | GTEx | DepMap | Descartes | 0.00 | 0.05 |
| SLC24A3 | 0.0000134 | 3194 | GTEx | DepMap | Descartes | 0.00 | 0.07 |
| FLI1 | 0.0000115 | 3249 | GTEx | DepMap | Descartes | 0.18 | 7.54 |
| ANGPT1 | 0.0000060 | 3402 | GTEx | DepMap | Descartes | 0.03 | 1.07 |
| RAB27B | 0.0000054 | 3426 | GTEx | DepMap | Descartes | 0.02 | 0.59 |
| GSN | -0.0000017 | 3672 | GTEx | DepMap | Descartes | 2.87 | 79.94 |
| GP1BA | -0.0000023 | 3694 | GTEx | DepMap | Descartes | 0.01 | 1.18 |
| STOM | -0.0000199 | 4450 | GTEx | DepMap | Descartes | 0.72 | 40.71 |
| TRPC6 | -0.0000215 | 4527 | GTEx | DepMap | Descartes | 0.02 | 0.69 |
| PDE3A | -0.0000317 | 4951 | GTEx | DepMap | Descartes | 0.04 | 0.95 |
| MYLK | -0.0000376 | 5213 | GTEx | DepMap | Descartes | 0.22 | 2.79 |
| MED12L | -0.0000403 | 5326 | GTEx | DepMap | Descartes | 0.01 | 0.15 |
| LTBP1 | -0.0000418 | 5386 | GTEx | DepMap | Descartes | 0.08 | 1.86 |
| DOK6 | -0.0000656 | 6416 | GTEx | DepMap | Descartes | 0.04 | 0.67 |
| VCL | -0.0000671 | 6487 | GTEx | DepMap | Descartes | 0.21 | 3.93 |
Descartes Lyphoid markers
Top 50 marker genes of Lyphoid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/lymphoid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 7.08e-01
Mean rank of genes in gene set: 6580.05
Median rank of genes in gene set: 7225
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ARID5B | 0.0008016 | 450 | GTEx | DepMap | Descartes | 0.75 | 19.52 |
| PLEKHA2 | 0.0005820 | 616 | GTEx | DepMap | Descartes | 0.29 | 11.46 |
| RCSD1 | 0.0001916 | 1352 | GTEx | DepMap | Descartes | 0.37 | 15.80 |
| ITPKB | 0.0001757 | 1434 | GTEx | DepMap | Descartes | 0.08 | 3.08 |
| DOCK10 | 0.0001442 | 1619 | GTEx | DepMap | Descartes | 0.37 | 11.35 |
| PRKCH | 0.0001266 | 1719 | GTEx | DepMap | Descartes | 0.14 | 9.92 |
| ANKRD44 | 0.0001216 | 1764 | GTEx | DepMap | Descartes | 0.29 | 8.38 |
| MCTP2 | 0.0001017 | 1941 | GTEx | DepMap | Descartes | 0.05 | 2.80 |
| ARHGAP15 | 0.0000961 | 1996 | GTEx | DepMap | Descartes | 0.40 | 32.36 |
| PTPRC | 0.0000933 | 2018 | GTEx | DepMap | Descartes | 2.05 | 87.02 |
| CCL5 | 0.0000896 | 2060 | GTEx | DepMap | Descartes | 0.91 | 178.86 |
| CELF2 | 0.0000802 | 2138 | GTEx | DepMap | Descartes | 0.78 | 21.53 |
| PITPNC1 | 0.0000697 | 2233 | GTEx | DepMap | Descartes | 0.22 | 7.00 |
| WIPF1 | 0.0000419 | 2588 | GTEx | DepMap | Descartes | 0.80 | 37.90 |
| LEF1 | 0.0000027 | 3524 | GTEx | DepMap | Descartes | 0.12 | 5.98 |
| PDE3B | -0.0000172 | 4328 | GTEx | DepMap | Descartes | 0.12 | 4.62 |
| FOXP1 | -0.0000238 | 4645 | GTEx | DepMap | Descartes | 0.70 | 14.93 |
| SORL1 | -0.0000363 | 5161 | GTEx | DepMap | Descartes | 0.42 | 8.66 |
| NCALD | -0.0000410 | 5353 | GTEx | DepMap | Descartes | 0.07 | 3.67 |
| EVL | -0.0000432 | 5445 | GTEx | DepMap | Descartes | 1.36 | 75.97 |
| STK39 | -0.0000717 | 6657 | GTEx | DepMap | Descartes | 0.14 | 7.02 |
| BACH2 | -0.0000984 | 7793 | GTEx | DepMap | Descartes | 0.08 | 1.89 |
| BCL2 | -0.0001024 | 7961 | GTEx | DepMap | Descartes | 0.29 | 9.06 |
| SP100 | -0.0001106 | 8258 | GTEx | DepMap | Descartes | 0.68 | 26.85 |
| MBNL1 | -0.0001240 | 8730 | GTEx | DepMap | Descartes | 0.79 | 26.12 |
| RAP1GAP2 | -0.0001459 | 9369 | GTEx | DepMap | Descartes | 0.12 | 2.90 |
| TOX | -0.0001537 | 9583 | GTEx | DepMap | Descartes | 0.09 | 5.04 |
| ABLIM1 | -0.0001580 | 9682 | GTEx | DepMap | Descartes | 0.14 | 3.92 |
| LCP1 | -0.0001589 | 9704 | GTEx | DepMap | Descartes | 1.66 | 83.27 |
| SAMD3 | -0.0001681 | 9936 | GTEx | DepMap | Descartes | 0.02 | 1.28 |
Below shows the ranks on this GEP for immune marker gene lists curated by CellTypist (https://www.celltypist.org/encyclopedia).
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Macrophages: Macrophages (model markers)
specialised mononuclear phagocytic cells which recognise:
Wilcoxon ranksum test P-value for gene set overrepresentation: 3.74e-04
Mean rank of genes in gene set: 169.25
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CD5L | 0.0057570 | 13 | GTEx | DepMap | Descartes | 1.32 | 105.92 |
| APOE | 0.0022957 | 115 | GTEx | DepMap | Descartes | 104.15 | 15735.56 |
| FTH1 | 0.0018445 | 153 | GTEx | DepMap | Descartes | 141.72 | 22732.38 |
| APOC1 | 0.0008809 | 396 | GTEx | DepMap | Descartes | 39.63 | 10013.99 |
Monocytes: Monocytes (model markers)
myeloid mononuclear recirculating leukocytes that are capable of differentiating into macrophages and myeloid lineage dendritic cells:
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.44e-03
Mean rank of genes in gene set: 386.67
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| SAT1 | 0.0049181 | 22 | GTEx | DepMap | Descartes | 37.79 | 6868.21 |
| NEAT1 | 0.0046948 | 26 | GTEx | DepMap | Descartes | 13.43 | 129.48 |
| TYROBP | 0.0002605 | 1112 | GTEx | DepMap | Descartes | 18.71 | 5956.44 |
Macrophages: Kupffer cells (model markers)
resident macrophages in liver under non-inflammatory conditions which line the hepatic sinusoids and are involved in erythrocyte clearance:
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.83e-03
Mean rank of genes in gene set: 1264
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CD5L | 0.0057570 | 13 | GTEx | DepMap | Descartes | 1.32 | 105.92 |
| EGR2 | 0.0018961 | 148 | GTEx | DepMap | Descartes | 1.04 | 73.09 |
| ARRDC3 | 0.0001002 | 1958 | GTEx | DepMap | Descartes | 0.76 | 40.79 |
| PENK | 0.0000236 | 2937 | GTEx | DepMap | Descartes | 0.05 | 5.61 |