Program: 30. Unclear.
Program description and justification of annotation generated by GPT5: Ribosome/Translation and ER co-translational targeting (MYC/MYCN-associated), ubiquitous with strongest activation in adrenergic neuroblastoma cells.
Submit a comment on this gene expression program's interpretation: CLICK
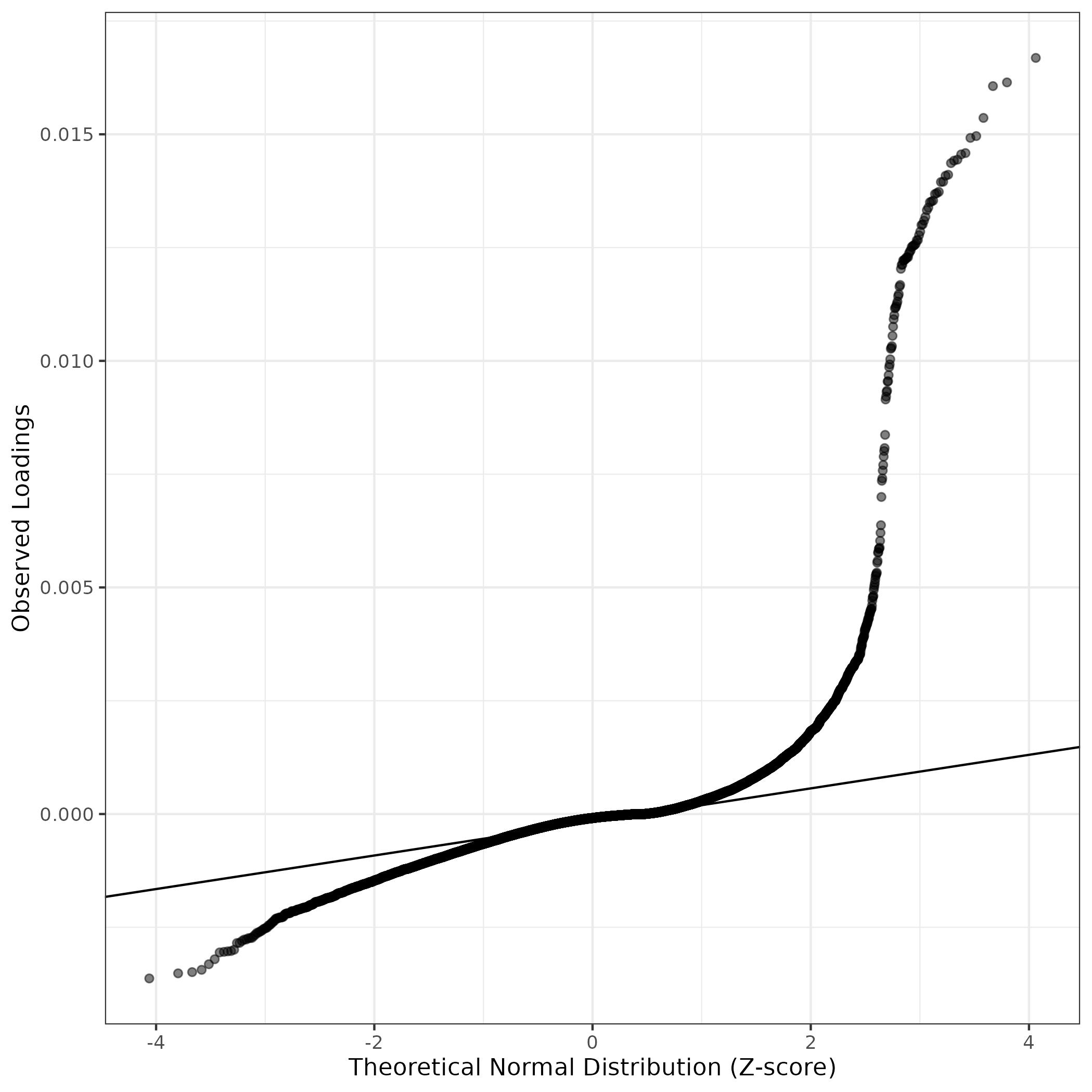
QQ-plot of gene loadings, averaged over both independent splits of the data
This plot highlights the relative contribution of each gene to the GEP
Top genes driving this program.
Note: Decartes website is buggy, try refreshing. Also, Decartes fetal adrenal data have been collected at specific time points (89-122 days), all possible cell types of interest may not be represented, do not overinterpret.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
| Gene | Loading | Gene.Name | GTEx | DepMap | Descartes | Mean.Counts | Mean.Tpm | |
|---|---|---|---|---|---|---|---|---|
| 1 | RPL18A | 0.0166859 | ribosomal protein L18a | GTEx | DepMap | Descartes | 6.92 | 3068.95 |
| 2 | RPL13 | 0.0161454 | ribosomal protein L13 | GTEx | DepMap | Descartes | 18.00 | 2459.51 |
| 3 | RPS2 | 0.0160635 | ribosomal protein S2 | GTEx | DepMap | Descartes | 10.51 | 4193.09 |
| 4 | RPL10 | 0.0153615 | ribosomal protein L10 | GTEx | DepMap | Descartes | 10.55 | 2725.06 |
| 5 | RPL32 | 0.0149610 | ribosomal protein L32 | GTEx | DepMap | Descartes | 8.75 | 2596.49 |
| 6 | RPS18 | 0.0149211 | ribosomal protein S18 | GTEx | DepMap | Descartes | 15.68 | 8546.52 |
| 7 | RPS23 | 0.0145858 | ribosomal protein S23 | GTEx | DepMap | Descartes | 9.47 | 1802.83 |
| 8 | RPS15 | 0.0145565 | ribosomal protein S15 | GTEx | DepMap | Descartes | 10.13 | 4004.58 |
| 9 | RPS19 | 0.0144405 | ribosomal protein S19 | GTEx | DepMap | Descartes | 8.95 | 2557.93 |
| 10 | RPS27 | 0.0144188 | ribosomal protein S27 | GTEx | DepMap | Descartes | 6.70 | 6659.67 |
| 11 | RPS3A | 0.0143640 | ribosomal protein S3A | GTEx | DepMap | Descartes | 8.40 | 3517.66 |
| 12 | RPS24 | 0.0141077 | ribosomal protein S24 | GTEx | DepMap | Descartes | 13.06 | 2503.22 |
| 13 | RPS3 | 0.0140852 | ribosomal protein S3 | GTEx | DepMap | Descartes | 8.25 | 2432.67 |
| 14 | RPL36 | 0.0139532 | ribosomal protein L36 | GTEx | DepMap | Descartes | 8.16 | 5540.31 |
| 15 | RPS14 | 0.0139471 | ribosomal protein S14 | GTEx | DepMap | Descartes | 8.27 | 1598.89 |
| 16 | RPS27A | 0.0137299 | ribosomal protein S27a | GTEx | DepMap | Descartes | 8.61 | 4415.76 |
| 17 | RPL35A | 0.0137022 | ribosomal protein L35a | GTEx | DepMap | Descartes | 5.85 | 2921.88 |
| 18 | RPLP1 | 0.0136827 | ribosomal protein lateral stalk subunit P1 | GTEx | DepMap | Descartes | 11.89 | 3629.98 |
| 19 | RPS4X | 0.0135370 | ribosomal protein S4 X-linked | GTEx | DepMap | Descartes | 6.56 | 2122.64 |
| 20 | RPL18 | 0.0135158 | ribosomal protein L18 | GTEx | DepMap | Descartes | 6.87 | 1064.24 |
| 21 | RPL29 | 0.0135010 | ribosomal protein L29 | GTEx | DepMap | Descartes | 6.58 | 4402.35 |
| 22 | RPL3 | 0.0133817 | ribosomal protein L3 | GTEx | DepMap | Descartes | 9.42 | 2618.05 |
| 23 | RPL7A | 0.0133248 | ribosomal protein L7a | GTEx | DepMap | Descartes | 6.93 | 3447.70 |
| 24 | RPS7 | 0.0131780 | ribosomal protein S7 | GTEx | DepMap | Descartes | 8.82 | 985.96 |
| 25 | RPS28 | 0.0130954 | ribosomal protein S28 | GTEx | DepMap | Descartes | 5.06 | 1950.01 |
| 26 | RPL24 | 0.0130127 | ribosomal protein L24 | GTEx | DepMap | Descartes | 6.15 | 2052.29 |
| 27 | RPL30 | 0.0129978 | ribosomal protein L30 | GTEx | DepMap | Descartes | 7.73 | 4239.91 |
| 28 | RPS12 | 0.0128531 | ribosomal protein S12 | GTEx | DepMap | Descartes | 5.93 | 5674.27 |
| 29 | RPL34 | 0.0127722 | ribosomal protein L34 | GTEx | DepMap | Descartes | 7.68 | 1826.40 |
| 30 | RPLP0 | 0.0126641 | ribosomal protein lateral stalk subunit P0 | GTEx | DepMap | Descartes | 6.42 | 1504.56 |
| 31 | RPL37A | 0.0126633 | ribosomal protein L37a | GTEx | DepMap | Descartes | 9.86 | 1993.94 |
| 32 | RPL13A | 0.0125852 | ribosomal protein L13a | GTEx | DepMap | Descartes | 7.08 | 1915.49 |
| 33 | RPL19 | 0.0125551 | ribosomal protein L19 | GTEx | DepMap | Descartes | 9.87 | 4886.10 |
| 34 | RPSA | 0.0125436 | ribosomal protein SA | GTEx | DepMap | Descartes | 6.36 | 1789.86 |
| 35 | RPS15A | 0.0125356 | ribosomal protein S15a | GTEx | DepMap | Descartes | 4.12 | 692.64 |
| 36 | RPS6 | 0.0125108 | ribosomal protein S6 | GTEx | DepMap | Descartes | 5.97 | 2720.72 |
| 37 | RPL15 | 0.0124386 | ribosomal protein L15 | GTEx | DepMap | Descartes | 7.02 | 1290.05 |
| 38 | RPS13 | 0.0124109 | ribosomal protein S13 | GTEx | DepMap | Descartes | 5.24 | 4271.61 |
| 39 | RACK1 | 0.0123656 | receptor for activated C kinase 1 | GTEx | DepMap | Descartes | 4.71 | NA |
| 40 | RPL41 | 0.0122859 | ribosomal protein L41 | GTEx | DepMap | Descartes | 6.27 | 5388.55 |
| 41 | RPL8 | 0.0122837 | ribosomal protein L8 | GTEx | DepMap | Descartes | 8.60 | 4927.99 |
| 42 | RPL12 | 0.0122804 | ribosomal protein L12 | GTEx | DepMap | Descartes | 4.56 | 1281.90 |
| 43 | RPL6 | 0.0122444 | ribosomal protein L6 | GTEx | DepMap | Descartes | 6.91 | 2127.11 |
| 44 | RPS5 | 0.0122322 | ribosomal protein S5 | GTEx | DepMap | Descartes | 4.52 | 1526.89 |
| 45 | RPL14 | 0.0122169 | ribosomal protein L14 | GTEx | DepMap | Descartes | 5.92 | 489.17 |
| 46 | RPL37 | 0.0122168 | ribosomal protein L37 | GTEx | DepMap | Descartes | 6.59 | 545.65 |
| 47 | RPS16 | 0.0121238 | ribosomal protein S16 | GTEx | DepMap | Descartes | 4.67 | 1207.33 |
| 48 | EEF1A1 | 0.0121184 | eukaryotic translation elongation factor 1 alpha 1 | GTEx | DepMap | Descartes | 7.90 | 1059.34 |
| 49 | RPL11 | 0.0120315 | ribosomal protein L11 | GTEx | DepMap | Descartes | 8.85 | 3384.12 |
| 50 | RPL5 | 0.0116748 | ribosomal protein L5 | GTEx | DepMap | Descartes | 5.88 | 2956.87 |
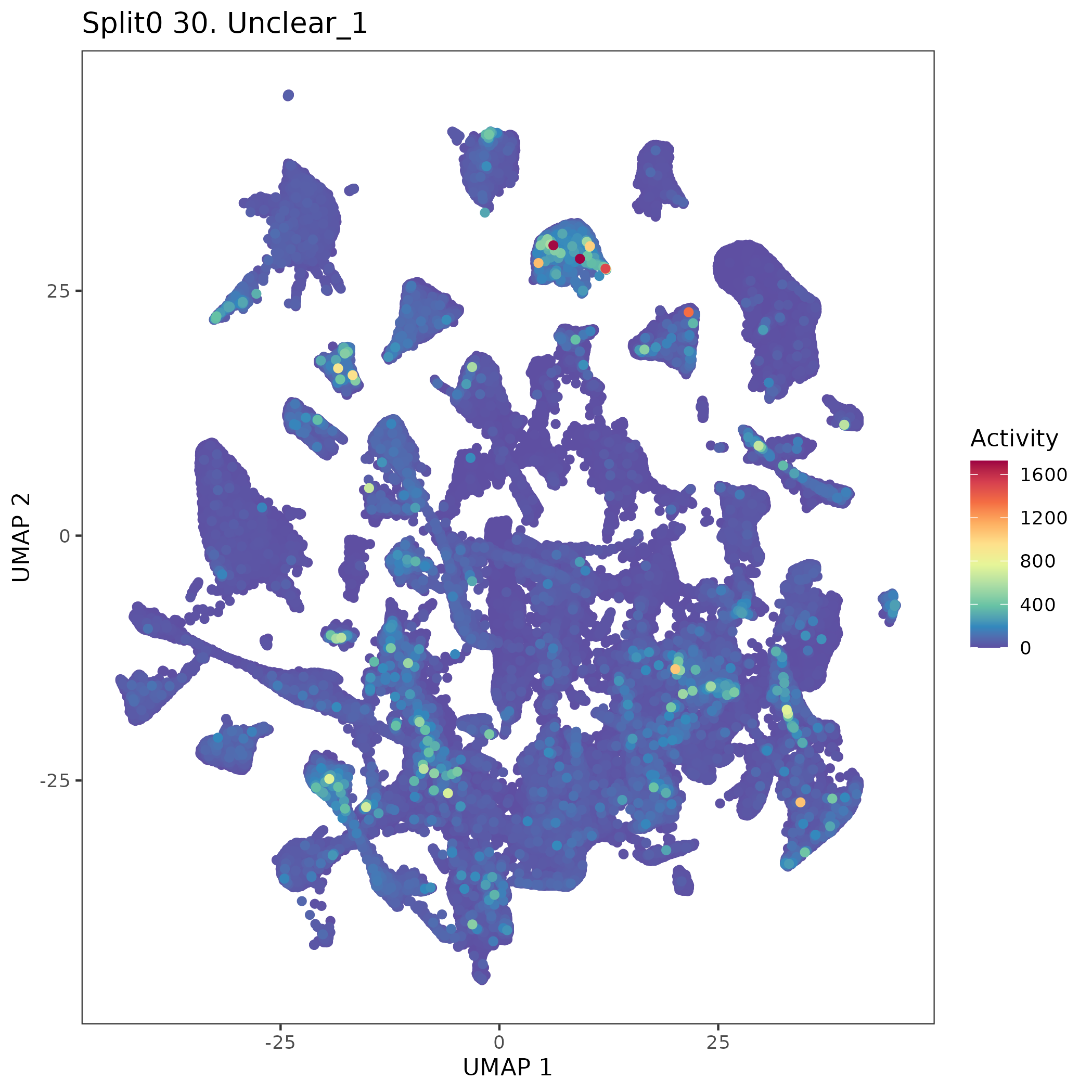
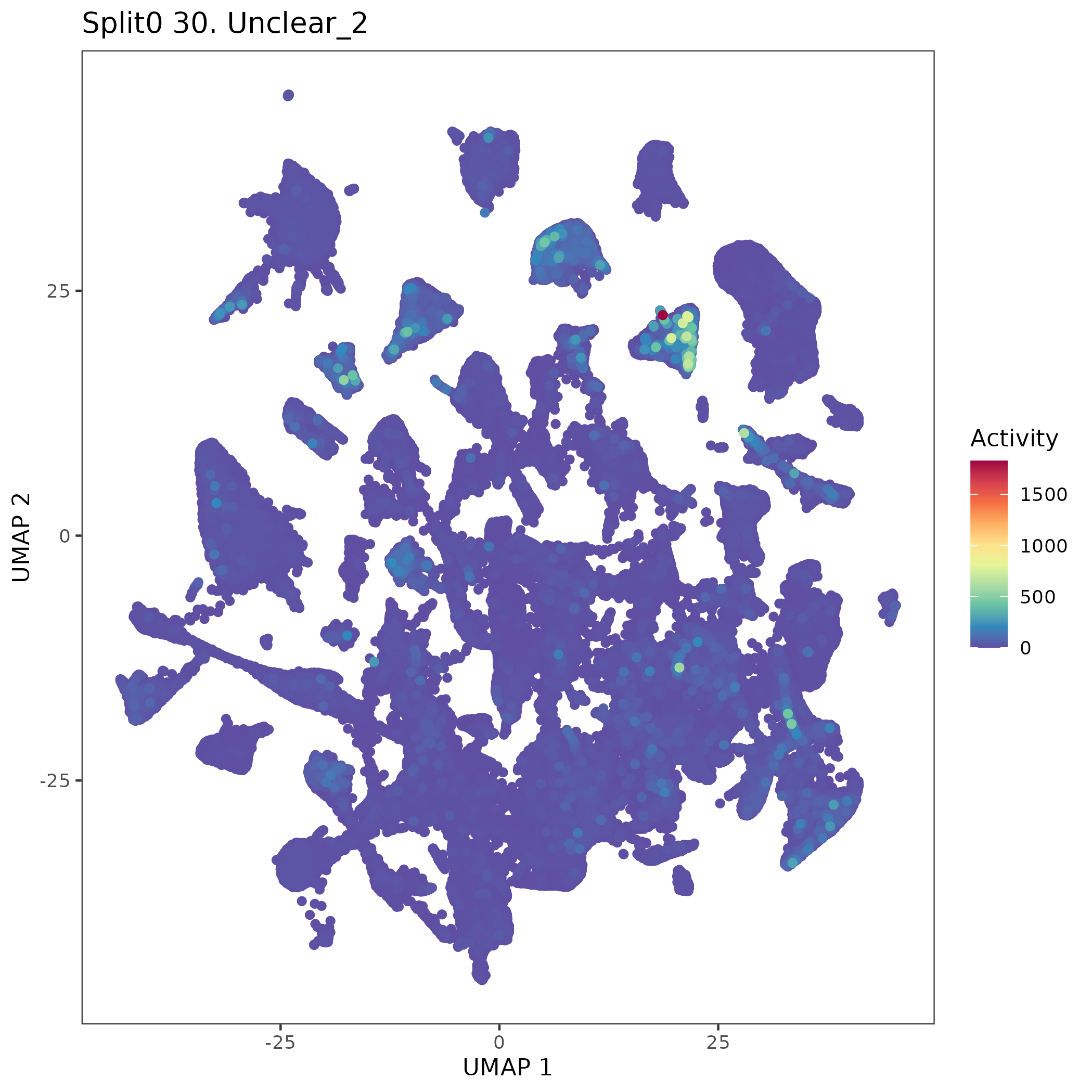
UMAP plots showing activity of gene expression program identified in GEP 30. Unclear:
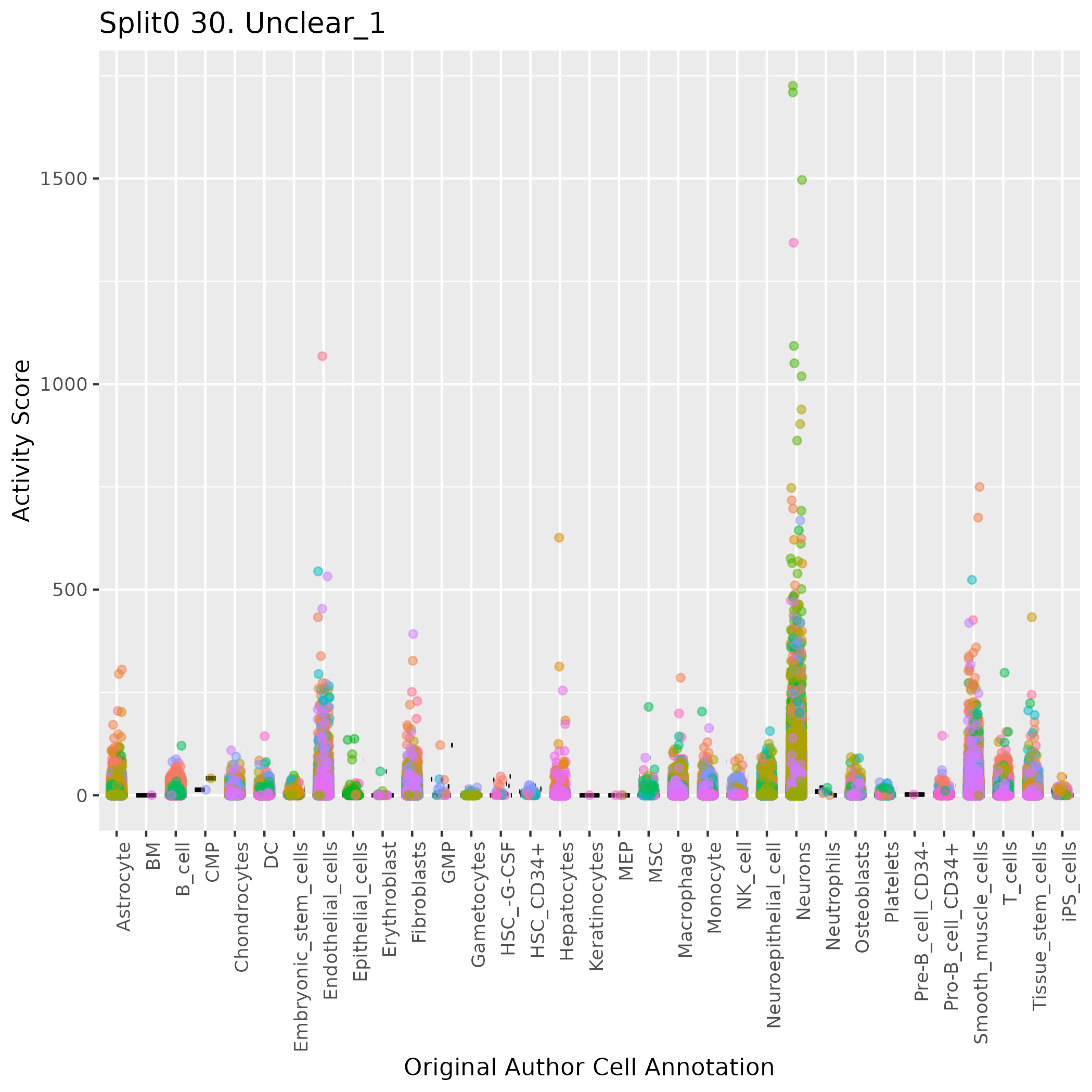
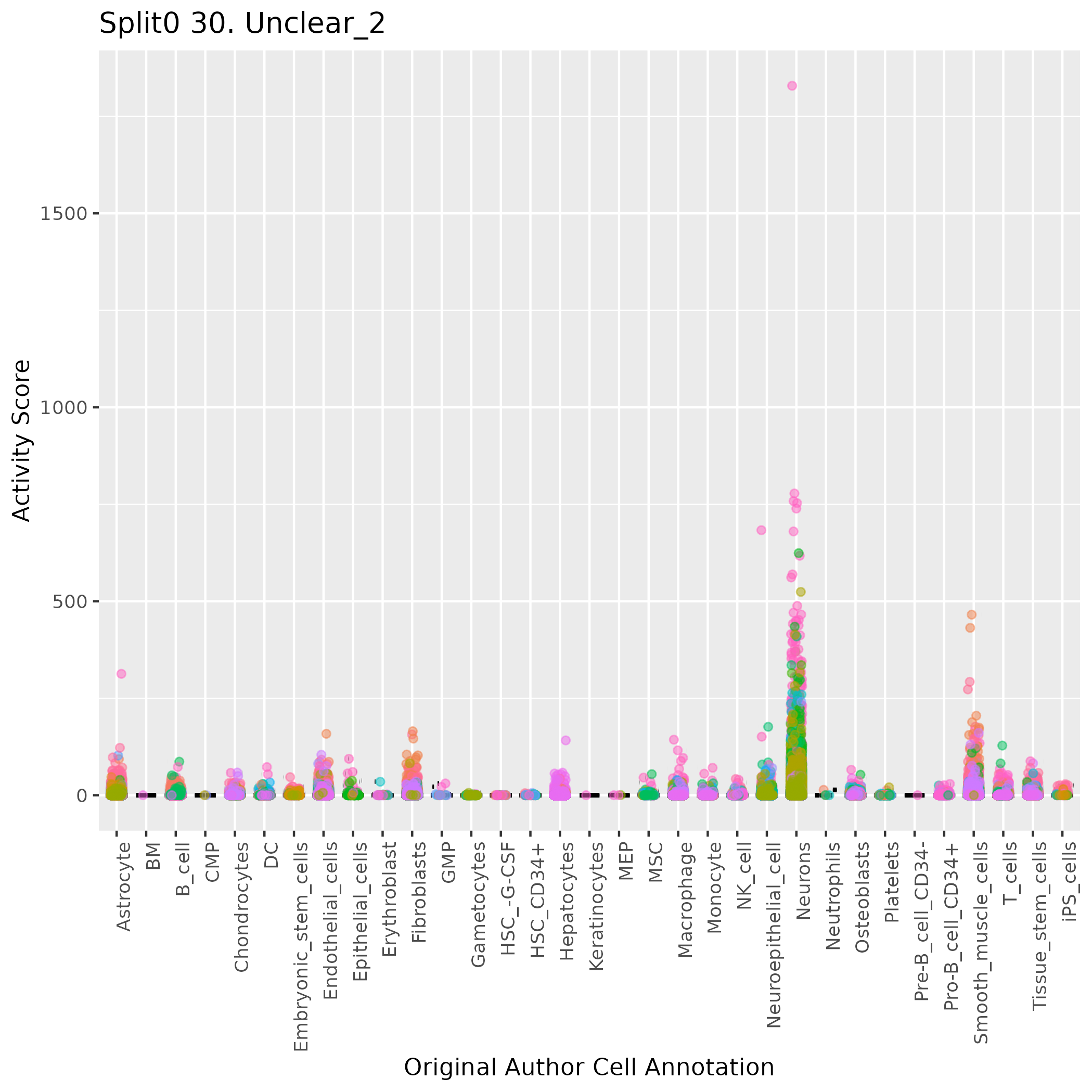
Boxlot showing activity of gene expression program identified in GEP 30. Unclear:
Gene set Enrichments for this program, caculated from top 50 genes
mSigDB Cell Types Gene Set:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| RUBENSTEIN_SKELETAL_MUSCLE_T_CELLS | 6.16e-107 | Inf | 2100.51 | 4.13e-104 | 4.13e-104 | 50RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, EEF1A1, RPL11, RPL5 |
181 |
| RUBENSTEIN_SKELETAL_MUSCLE_SATELLITE_CELLS | 5.95e-94 | Inf | 1050.26 | 7.99e-92 | 4.00e-91 | 50RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, EEF1A1, RPL11, RPL5 |
310 |
| BUSSLINGER_GASTRIC_PPP1R1B_POSITIVE_CELLS | 1.12e-95 | 1976.54 | 791.12 | 2.50e-93 | 7.50e-93 | 44RPL18A, RPL13, RPS2, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS28, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPS5, RPL14, RPL37, RPS16, EEF1A1, RPL11, RPL5 |
121 |
| RUBENSTEIN_SKELETAL_MUSCLE_B_CELLS | 1.34e-96 | 2341.84 | 760.13 | 4.50e-94 | 9.01e-94 | 47RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL30, RPS12, RPL34, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, EEF1A1, RPL11, RPL5 |
177 |
| BUSSLINGER_DUODENAL_TRANSIT_AMPLIFYING_CELLS | 1.83e-94 | 2087.76 | 690.91 | 3.07e-92 | 1.23e-91 | 47RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, EEF1A1, RPL11, RPL5 |
194 |
| TRAVAGLINI_LUNG_CD4_NAIVE_T_CELL | 1.18e-93 | 1735.10 | 671.22 | 1.32e-91 | 7.90e-91 | 44RPL18A, RPL13, RPS2, RPL10, RPL32, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPL18, RPL29, RPL3, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPS15A, RPS6, RPL15, RPS13, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, EEF1A1, RPL11, RPL5 |
132 |
| BUSSLINGER_DUODENAL_DIFFERENTIATING_STEM_CELLS | 9.99e-91 | 3775.62 | 668.68 | 9.58e-89 | 6.71e-88 | 49RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, EEF1A1, RPL11, RPL5 |
305 |
| BUSSLINGER_GASTRIC_LYZ_POSITIVE_CELLS | 4.16e-88 | 1360.97 | 600.92 | 3.49e-86 | 2.79e-85 | 41RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS27, RPS3A, RPS3, RPL36, RPS14, RPL35A, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL19, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL12, RPL6, RPS5, RPL14, RPL37, EEF1A1, RPL11, RPL5 |
111 |
| TRAVAGLINI_LUNG_BRONCHIAL_VESSEL_1_CELL | 4.81e-88 | 1329.18 | 478.71 | 3.58e-86 | 3.23e-85 | 46RPL18A, RPL13, RPS2, RPL10, RPL32, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
224 |
| BUSSLINGER_DUODENAL_STEM_CELLS | 6.08e-81 | 896.42 | 320.24 | 4.08e-79 | 4.08e-78 | 46RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPL37A, RPL13A, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, EEF1A1, RPL11, RPL5 |
311 |
| TRAVAGLINI_LUNG_CLUB_CELL | 5.44e-70 | 601.41 | 304.13 | 2.15e-68 | 3.65e-67 | 35RPL18A, RPL13, RPL10, RPL32, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPL35A, RPL18, RPL29, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPL37A, RPL13A, RPL19, RPS15A, RPS6, RPL15, RPS13, RPL12, RPS5, RPL37, RPS16, RPL11, RPL5 |
114 |
| FAN_OVARY_CL0_XBP1_SELK_HIGH_STROMAL_CELL | 3.94e-74 | 621.03 | 302.39 | 2.03e-72 | 2.64e-71 | 38RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL3, RPL24, RPL30, RPS12, RPL34, RPL13A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL11, RPL5 |
143 |
| LAKE_ADULT_KIDNEY_C7_PROXIMAL_TUBULE_EPITHELIAL_CELLS_S3 | 1.06e-69 | 531.01 | 271.26 | 3.96e-68 | 7.12e-67 | 36RPL13, RPL32, RPS18, RPS23, RPS15, RPS19, RPS3A, RPS24, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL3, RPL7A, RPS7, RPS28, RPL24, RPS12, RPL34, RPL37A, RPL13A, RPL19, RPSA, RPS6, RPS13, RPL8, RPL12, RPL6, RPL14, RPL37, RPS16, EEF1A1, RPL11, RPL5 |
135 |
| LAKE_ADULT_KIDNEY_C19_COLLECTING_DUCT_INTERCALATED_CELLS_TYPE_A_MEDULLA | 4.09e-77 | 657.54 | 258.22 | 2.49e-75 | 2.74e-74 | 45RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPS16, EEF1A1, RPL11, RPL5 |
326 |
| LAKE_ADULT_KIDNEY_C8_DECENDING_THIN_LIMB | 7.17e-75 | 536.14 | 236.51 | 4.01e-73 | 4.81e-72 | 43RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS14, RPS27A, RPLP1, RPS4X, RPL18, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPS16, EEF1A1, RPL11, RPL5 |
278 |
| LAKE_ADULT_KIDNEY_C12_THICK_ASCENDING_LIMB | 6.45e-74 | 550.07 | 217.37 | 3.09e-72 | 4.33e-71 | 45RPL13, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, EEF1A1, RPL11, RPL5 |
381 |
| LAKE_ADULT_KIDNEY_C18_COLLECTING_DUCT_PRINCIPAL_CELLS_MEDULLA | 5.85e-72 | 454.36 | 199.45 | 2.62e-70 | 3.93e-69 | 43RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS14, RPS27A, RPLP1, RPS4X, RPL18, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPS16, EEF1A1, RPL11, RPL5 |
322 |
| LAKE_ADULT_KIDNEY_C9_THIN_ASCENDING_LIMB | 2.75e-70 | 412.25 | 194.25 | 1.15e-68 | 1.85e-67 | 41RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS3A, RPS24, RPS3, RPS14, RPS27A, RPLP1, RPS4X, RPL18, RPL3, RPL7A, RPS7, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS6, RPL15, RPS13, RACK1, RPL8, RPL12, RPL6, RPS5, RPL14, RPS16, EEF1A1, RPL11, RPL5 |
268 |
| TRAVAGLINI_LUNG_PROXIMAL_BASAL_CELL | 4.99e-69 | 548.55 | 176.67 | 1.76e-67 | 3.35e-66 | 47RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL30, RPS12, RPL34, RPLP0, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, EEF1A1, RPL11, RPL5 |
629 |
| LAKE_ADULT_KIDNEY_C10_THIN_ASCENDING_LIMB | 1.32e-67 | 342.85 | 159.69 | 4.42e-66 | 8.84e-65 | 42RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS14, RPS27A, RPLP1, RPS4X, RPL18, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPS16, EEF1A1, RPL11, RPL5 |
353 |
Dowload full table
mSigDB Hallmark Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| HALLMARK_MYC_TARGETS_V1 | 3.99e-11 | 27.00 | 11.87 | 1.99e-09 | 1.99e-09 | 10RPS2, RPS3, RPL18, RPL34, RPLP0, RPS6, RACK1, RPL6, RPS5, RPL14 |
200 |
| HALLMARK_P53_PATHWAY | 1.36e-03 | 9.11 | 2.36 | 3.39e-02 | 6.78e-02 | 4RPL36, RPL18, RPS12, RACK1 |
200 |
| HALLMARK_ALLOGRAFT_REJECTION | 8.36e-02 | 4.32 | 0.51 | 1.00e+00 | 1.00e+00 | 2RPS19, RPS3A |
200 |
| HALLMARK_UNFOLDED_PROTEIN_RESPONSE | 2.40e-01 | 3.74 | 0.09 | 1.00e+00 | 1.00e+00 | 1RPS14 |
113 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
200 |
| HALLMARK_HYPOXIA | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
200 |
| HALLMARK_CHOLESTEROL_HOMEOSTASIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
74 |
| HALLMARK_MITOTIC_SPINDLE | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
199 |
| HALLMARK_WNT_BETA_CATENIN_SIGNALING | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
42 |
| HALLMARK_TGF_BETA_SIGNALING | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
54 |
| HALLMARK_IL6_JAK_STAT3_SIGNALING | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
87 |
| HALLMARK_DNA_REPAIR | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
150 |
| HALLMARK_G2M_CHECKPOINT | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
200 |
| HALLMARK_APOPTOSIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
161 |
| HALLMARK_NOTCH_SIGNALING | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
32 |
| HALLMARK_ADIPOGENESIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
200 |
| HALLMARK_ESTROGEN_RESPONSE_EARLY | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
200 |
| HALLMARK_ESTROGEN_RESPONSE_LATE | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
200 |
| HALLMARK_ANDROGEN_RESPONSE | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
100 |
| HALLMARK_MYOGENESIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
200 |
Dowload full table
KEGG Pathways:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| KEGG_RIBOSOME | 1.70e-114 | 9969.60 | 2409.41 | 3.17e-112 | 3.17e-112 | 47RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
88 |
| KEGG_MTOR_SIGNALING_PATHWAY | 1.19e-01 | 8.21 | 0.20 | 1.00e+00 | 1.00e+00 | 1RPS6 |
52 |
| KEGG_INSULIN_SIGNALING_PATHWAY | 2.83e-01 | 3.08 | 0.08 | 1.00e+00 | 1.00e+00 | 1RPS6 |
137 |
| KEGG_N_GLYCAN_BIOSYNTHESIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
46 |
| KEGG_OTHER_GLYCAN_DEGRADATION | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
16 |
| KEGG_O_GLYCAN_BIOSYNTHESIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
30 |
| KEGG_GLYCOSAMINOGLYCAN_DEGRADATION | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
21 |
| KEGG_GLYCOSAMINOGLYCAN_BIOSYNTHESIS_KERATAN_SULFATE | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
15 |
| KEGG_GLYCEROLIPID_METABOLISM | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
49 |
| KEGG_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI_ANCHOR_BIOSYNTHESIS | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
25 |
| KEGG_GLYCEROPHOSPHOLIPID_METABOLISM | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
77 |
| KEGG_ETHER_LIPID_METABOLISM | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
33 |
| KEGG_ARACHIDONIC_ACID_METABOLISM | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
58 |
| KEGG_LINOLEIC_ACID_METABOLISM | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
29 |
| KEGG_ALPHA_LINOLENIC_ACID_METABOLISM | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
19 |
| KEGG_SPHINGOLIPID_METABOLISM | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
39 |
| KEGG_GLYCOSPHINGOLIPID_BIOSYNTHESIS_LACTO_AND_NEOLACTO_SERIES | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
26 |
| KEGG_GLYCOSPHINGOLIPID_BIOSYNTHESIS_GLOBO_SERIES | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
14 |
| KEGG_GLYCOSPHINGOLIPID_BIOSYNTHESIS_GANGLIO_SERIES | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
15 |
| KEGG_RIBOFLAVIN_METABOLISM | 1.00e+00 | 0.00 | 0.00 | 1.00e+00 | 1.00e+00 | 0 |
16 |
Dowload full table
CHR Positional Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| chr3p22 | 6.01e-02 | 5.25 | 0.61 | 1.00e+00 | 1.00e+00 | 2RPSA, RPL14 |
165 |
| chr19q13 | 1.92e-01 | 1.97 | 0.61 | 1.00e+00 | 1.00e+00 | 5RPS19, RPL18, RPL13A, RPS5, RPS16 |
1165 |
| chr19p13 | 1.06e-01 | 2.32 | 0.61 | 1.00e+00 | 1.00e+00 | 4RPL18A, RPS15, RPL36, RPS28 |
773 |
| chr12q24 | 2.38e-01 | 2.21 | 0.26 | 1.00e+00 | 1.00e+00 | 2RPLP0, RPL6 |
390 |
| chr3q12 | 1.06e-01 | 9.31 | 0.23 | 1.00e+00 | 1.00e+00 | 1RPL24 |
46 |
| chr6q13 | 1.27e-01 | 7.62 | 0.19 | 1.00e+00 | 1.00e+00 | 1EEF1A1 |
56 |
| chr15q23 | 1.46e-01 | 6.55 | 0.16 | 1.00e+00 | 1.00e+00 | 1RPLP1 |
65 |
| chr4q25 | 1.90e-01 | 4.87 | 0.12 | 1.00e+00 | 1.00e+00 | 1RPL34 |
87 |
| chr3p24 | 2.14e-01 | 4.28 | 0.11 | 1.00e+00 | 1.00e+00 | 1RPL15 |
99 |
| chr6q23 | 2.27e-01 | 3.99 | 0.10 | 1.00e+00 | 1.00e+00 | 1RPS12 |
106 |
| chr5q33 | 2.32e-01 | 3.88 | 0.10 | 1.00e+00 | 1.00e+00 | 1RPS14 |
109 |
| chr2p16 | 2.42e-01 | 3.71 | 0.09 | 1.00e+00 | 1.00e+00 | 1RPS27A |
114 |
| chr2p25 | 2.47e-01 | 3.61 | 0.09 | 1.00e+00 | 1.00e+00 | 1RPS7 |
117 |
| chr3q29 | 2.56e-01 | 3.46 | 0.09 | 1.00e+00 | 1.00e+00 | 1RPL35A |
122 |
| chr2q35 | 2.63e-01 | 3.35 | 0.08 | 1.00e+00 | 1.00e+00 | 1RPL37A |
126 |
| chr5p13 | 2.67e-01 | 3.30 | 0.08 | 1.00e+00 | 1.00e+00 | 1RPL37 |
128 |
| chr1p22 | 2.69e-01 | 3.27 | 0.08 | 1.00e+00 | 1.00e+00 | 1RPL5 |
129 |
| chr16q24 | 2.70e-01 | 3.25 | 0.08 | 1.00e+00 | 1.00e+00 | 1RPL13 |
130 |
| chr5q14 | 2.70e-01 | 3.25 | 0.08 | 1.00e+00 | 1.00e+00 | 1RPS23 |
130 |
| chr9q33 | 2.72e-01 | 3.22 | 0.08 | 1.00e+00 | 1.00e+00 | 1RPL12 |
131 |
Dowload full table
Transcription Factor Targets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| ZZZ3_TARGET_GENES | 1.96e-20 | 74.52 | 36.06 | 5.55e-18 | 2.22e-17 | 14RPS2, RPS18, RPS23, RPS3A, RPS3, RPS14, RPL35A, RPS7, RPL24, RPL37A, RPL41, RPL8, RPL6, RPL37 |
121 |
| GTF2A2_TARGET_GENES | 6.33e-29 | 44.88 | 24.57 | 3.59e-26 | 7.17e-26 | 26RPL13, RPS2, RPL10, RPS18, RPS27, RPS3A, RPS24, RPS3, RPL36, RPLP1, RPL18, RPL29, RPL3, RPS7, RPL24, RPS12, RPLP0, RPL37A, RPSA, RPS6, RPL15, RPL8, RPL12, RPS5, EEF1A1, RPL11 |
522 |
| SNRNP70_TARGET_GENES | 3.45e-32 | 40.88 | 22.03 | 3.90e-29 | 3.90e-29 | 33RPL13, RPS2, RPL10, RPL32, RPS23, RPS15, RPS19, RPS24, RPS3, RPS14, RPL35A, RPL18, RPL3, RPL7A, RPL30, RPLP0, RPL13A, RPL19, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL37, RPS16, EEF1A1, RPL11, RPL5 |
1009 |
| TAF9B_TARGET_GENES | 6.85e-25 | 35.04 | 19.13 | 2.59e-22 | 7.76e-22 | 24RPL10, RPS15, RPS27, RPS24, RPS3, RPS14, RPL35A, RPS4X, RPL29, RPL3, RPL7A, RPS28, RPL34, RPL37A, RPL13A, RPS6, RACK1, RPL41, RPL12, RPL6, RPL14, EEF1A1, RPL11, RPL5 |
565 |
| HJURP_TARGET_GENES | 6.01e-07 | 73.99 | 18.01 | 2.30e-05 | 6.80e-04 | 4RPL3, RPS15A, RACK1, EEF1A1 |
28 |
| PER1_TARGET_GENES | 1.19e-06 | 61.45 | 15.09 | 4.21e-05 | 1.35e-03 | 4RPL3, RPL34, RPL12, EEF1A1 |
33 |
| FOXR2_TARGET_GENES | 2.55e-13 | 29.19 | 13.70 | 2.63e-11 | 2.89e-10 | 12RPL32, RPS18, RPS27, RPS14, RPL34, RPS6, RACK1, RPL8, RPL6, RPS5, RPS16, EEF1A1 |
234 |
| ZNF318_TARGET_GENES | 3.13e-17 | 24.22 | 12.70 | 5.06e-15 | 3.55e-14 | 18RPL13, RPL32, RPS18, RPS3A, RPS24, RPS3, RPS14, RPL29, RPL7A, RPS7, RPL34, RPL37A, RPS15A, RACK1, RPL41, RPL37, EEF1A1, RPL5 |
495 |
| FOXE1_TARGET_GENES | 1.44e-19 | 22.87 | 12.40 | 3.26e-17 | 1.63e-16 | 22RPL13, RPL32, RPS15, RPS27, RPS24, RPS3, RPS14, RPS4X, RPL18, RPL29, RPL3, RPS28, RPS12, RPLP0, RPS15A, RPS6, RPL15, RPL41, RPL12, RPL6, RPL11, RPL5 |
728 |
| PSMB5_TARGET_GENES | 2.52e-13 | 24.55 | 11.84 | 2.63e-11 | 2.85e-10 | 13RPS2, RPL10, RPL32, RPS27, RPLP1, RPL3, RPS28, RPS12, RPLP0, RPL13A, RPSA, RPL12, EEF1A1 |
307 |
| ZFHX3_TARGET_GENES | 4.04e-19 | 15.53 | 8.54 | 7.63e-17 | 4.58e-16 | 29RPL10, RPS18, RPS23, RPS27, RPS24, RPS3, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL3, RPL7A, RPL30, RPS12, RPL34, RPLP0, RPL13A, RPL19, RPS15A, RPS6, RPS13, RACK1, RPL8, RPS5, RPL37, EEF1A1, RPL5 |
1857 |
| GTF2E2_TARGET_GENES | 1.67e-10 | 16.26 | 7.67 | 1.35e-08 | 1.90e-07 | 12RPL10, RPL36, RPLP1, RPS28, RPS12, RPLP0, RPS6, RACK1, RPL41, RPL12, RPL37, EEF1A1 |
411 |
| RUVBL1_TARGET_GENES | 2.39e-07 | 19.32 | 7.23 | 1.12e-05 | 2.70e-04 | 7RPS14, RPL18, RPL24, RPS12, RPL41, EEF1A1, RPL11 |
180 |
| KAT2A_TARGET_GENES | 2.27e-14 | 13.10 | 7.08 | 2.85e-12 | 2.57e-11 | 21RPS2, RPS15, RPS3, RPL36, RPL35A, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS12, RPL34, RPL13A, RPS13, RACK1, RPL41, RPL8, RPL6, RPS16, RPL5 |
1157 |
| ADA2_TARGET_GENES | 3.39e-12 | 12.77 | 6.64 | 3.20e-10 | 3.84e-09 | 17RPL13, RPS2, RPL32, RPS18, RPS3A, RPS24, RPS3, RPS14, RPL18, RPL29, RPL3, RPS7, RPL8, RPL37, EEF1A1, RPL11, RPL5 |
846 |
| NFRKB_TARGET_GENES | 1.06e-14 | 11.31 | 6.22 | 1.49e-12 | 1.20e-11 | 25RPL32, RPS18, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPL35A, RPL29, RPL3, RPS7, RPL30, RPL34, RPL13A, RPS15A, RPS6, RPS13, RACK1, RPL41, RPL8, RPL6, RPL14, RPL37, RPL11 |
1843 |
| KAT5_TARGET_GENES | 1.19e-10 | 10.81 | 5.55 | 1.04e-08 | 1.35e-07 | 16RPL18A, RPL13, RPL10, RPL32, RPS23, RPS15, RPS24, RPL36, RPS14, RPL34, RPL37A, RPL19, RPSA, RPS6, EEF1A1, RPL5 |
910 |
| CHAF1B_TARGET_GENES | 3.41e-10 | 10.02 | 5.14 | 2.42e-08 | 3.86e-07 | 16RPS2, RPS27, RPL36, RPLP1, RPL18, RPL3, RPS7, RPL24, RPL30, RPS12, RPL13A, RPS15A, RPL12, RPS16, EEF1A1, RPL5 |
981 |
| SETD7_TARGET_GENES | 3.93e-10 | 9.92 | 5.09 | 2.62e-08 | 4.45e-07 | 16RPL13, RPS2, RPL10, RPS15, RPS19, RPS3A, RPS14, RPL3, RPL13A, RPS15A, RPS6, RPL15, RACK1, RPL14, EEF1A1, RPL5 |
991 |
| TET1_TARGET_GENES | 5.70e-09 | 9.50 | 4.71 | 3.40e-07 | 6.45e-06 | 14RPL18A, RPS19, RPS3A, RPS24, RPL35A, RPL29, RPL3, RPS7, RPS28, RPL24, RPS15A, RPS6, RPL12, RPL5 |
855 |
Dowload full table
GO Biological Processes:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GOBP_COTRANSLATIONAL_PROTEIN_TARGETING_TO_MEMBRANE | 7.88e-113 | 7827.50 | 2100.51 | 5.89e-109 | 5.89e-109 | 48RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
107 |
| GOBP_NUCLEAR_TRANSCRIBED_MRNA_CATABOLIC_PROCESS_NONSENSE_MEDIATED_DECAY | 9.43e-110 | 6515.45 | 1794.17 | 3.53e-106 | 7.06e-106 | 48RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
120 |
| GOBP_ESTABLISHMENT_OF_PROTEIN_LOCALIZATION_TO_ENDOPLASMIC_RETICULUM | 1.56e-109 | 6432.20 | 1777.46 | 3.89e-106 | 1.17e-105 | 48RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
121 |
| GOBP_PROTEIN_LOCALIZATION_TO_ENDOPLASMIC_RETICULUM | 3.25e-104 | 4731.23 | 1204.71 | 6.08e-101 | 2.43e-100 | 48RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
149 |
| GOBP_VIRAL_GENE_EXPRESSION | 2.48e-97 | 3227.55 | 873.99 | 3.09e-94 | 1.86e-93 | 48RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
198 |
| GOBP_TRANSLATIONAL_INITIATION | 6.16e-98 | 3335.96 | 859.01 | 9.22e-95 | 4.61e-94 | 48RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
193 |
| GOBP_NUCLEAR_TRANSCRIBED_MRNA_CATABOLIC_PROCESS | 3.57e-96 | 3030.49 | 810.98 | 3.34e-93 | 2.67e-92 | 48RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
208 |
| GOBP_PROTEIN_TARGETING_TO_MEMBRANE | 3.57e-96 | 3030.49 | 810.98 | 3.34e-93 | 2.67e-92 | 48RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
208 |
| GOBP_ESTABLISHMENT_OF_PROTEIN_LOCALIZATION_TO_MEMBRANE | 1.53e-87 | 3223.65 | 561.10 | 1.27e-84 | 1.15e-83 | 49RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
351 |
| GOBP_PEPTIDE_BIOSYNTHETIC_PROCESS | 2.02e-74 | Inf | 384.86 | 1.26e-71 | 1.51e-70 | 50RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, EEF1A1, RPL11, RPL5 |
740 |
| GOBP_RNA_CATABOLIC_PROCESS | 1.03e-80 | 1340.25 | 349.09 | 7.74e-78 | 7.74e-77 | 48RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
414 |
| GOBP_PROTEIN_TARGETING | 1.72e-79 | 1258.39 | 331.68 | 1.17e-76 | 1.28e-75 | 48RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
438 |
| GOBP_AMIDE_BIOSYNTHETIC_PROCESS | 9.37e-71 | Inf | 324.73 | 4.38e-68 | 7.01e-67 | 50RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, EEF1A1, RPL11, RPL5 |
877 |
| GOBP_PEPTIDE_METABOLIC_PROCESS | 3.96e-70 | Inf | 314.82 | 1.74e-67 | 2.97e-66 | 50RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, EEF1A1, RPL11, RPL5 |
903 |
| GOBP_PROTEIN_LOCALIZATION_TO_MEMBRANE | 9.40e-74 | 1629.24 | 283.36 | 5.41e-71 | 7.03e-70 | 49RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
658 |
| GOBP_ESTABLISHMENT_OF_PROTEIN_LOCALIZATION_TO_ORGANELLE | 1.23e-73 | 931.24 | 241.99 | 6.55e-71 | 9.17e-70 | 48RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
576 |
| GOBP_CELLULAR_AMIDE_METABOLIC_PROCESS | 2.30e-64 | Inf | 234.91 | 8.59e-62 | 1.72e-60 | 50RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, EEF1A1, RPL11, RPL5 |
1185 |
| GOBP_ORGANIC_CYCLIC_COMPOUND_CATABOLIC_PROCESS | 1.62e-71 | 835.23 | 219.96 | 8.09e-69 | 1.21e-67 | 48RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
637 |
| GOBP_MACROMOLECULE_CATABOLIC_PROCESS | 1.83e-60 | Inf | 195.37 | 5.96e-58 | 1.37e-56 | 50RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, EEF1A1, RPL11, RPL5 |
1429 |
| GOBP_BIOLOGICAL_PROCESS_INVOLVED_IN_SYMBIOTIC_INTERACTION | 5.59e-65 | 1053.13 | 181.28 | 2.32e-62 | 4.18e-61 | 49RPL18A, RPL13, RPS2, RPL10, RPL32, RPS18, RPS23, RPS15, RPS19, RPS27, RPS3A, RPS24, RPS3, RPL36, RPS14, RPS27A, RPL35A, RPLP1, RPS4X, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS28, RPL24, RPL30, RPS12, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
997 |
Dowload full table
Immunological Gene Sets:
| P-value | OR | Lower 95% CI | FDR | FWER | Genes Found | Gene Set Size | |
|---|---|---|---|---|---|---|---|
| GSE2405_0H_VS_24H_A_PHAGOCYTOPHILUM_STIM_NEUTROPHIL_UP | 2.28e-65 | 354.49 | 181.64 | 1.11e-61 | 1.11e-61 | 37RPL18A, RPL13, RPS2, RPL10, RPS18, RPS23, RPS19, RPS27, RPS3A, RPS3, RPL36, RPS14, RPS4X, RPL18, RPL3, RPL7A, RPS7, RPL24, RPL30, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RACK1, RPL41, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
200 |
| GSE2405_0H_VS_9H_A_PHAGOCYTOPHILUM_STIM_NEUTROPHIL_DN | 7.56e-63 | 320.65 | 165.71 | 1.84e-59 | 3.68e-59 | 36RPL18A, RPL13, RPS2, RPL10, RPS18, RPS23, RPS19, RPS27, RPS3A, RPS3, RPL36, RPS14, RPS4X, RPL18, RPL3, RPL7A, RPS7, RPL24, RPL30, RPL34, RPLP0, RPL37A, RPL13A, RPL19, RPSA, RPS15A, RPS6, RACK1, RPL12, RPL6, RPS5, RPL14, RPL37, RPS16, RPL11, RPL5 |
200 |
| GSE41978_ID2_KO_VS_ID2_KO_AND_BIM_KO_KLRG1_LOW_EFFECTOR_CD8_TCELL_DN | 3.54e-44 | 151.48 | 81.77 | 5.75e-41 | 1.73e-40 | 28RPL13, RPL10, RPS18, RPS23, RPS3A, RPS3, RPS14, RPLP1, RPS4X, RPL18, RPL3, RPS7, RPL24, RPL34, RPLP0, RPL37A, RPL19, RPSA, RPS15A, RPS6, RACK1, RPL41, RPL12, RPL6, RPS5, RPS16, RPL11, RPL5 |
200 |
| GSE41978_KLRG1_HIGH_VS_LOW_EFFECTOR_CD8_TCELL_DN | 5.13e-42 | 138.95 | 75.21 | 6.25e-39 | 2.50e-38 | 27RPS2, RPL10, RPS18, RPS23, RPS19, RPS3A, RPS3, RPS14, RPLP1, RPS4X, RPL18, RPL3, RPL7A, RPS7, RPL24, RPL34, RPLP0, RPL19, RPS15A, RPS6, RPL12, RPL6, RPS5, RPL37, RPS16, RPL11, RPL5 |
200 |
| GSE22886_NAIVE_BCELL_VS_NEUTROPHIL_UP | 5.95e-40 | 128.59 | 69.37 | 5.80e-37 | 2.90e-36 | 26RPL10, RPS18, RPS15, RPS19, RPS27, RPS24, RPS14, RPS27A, RPL35A, RPLP1, RPL7A, RPS7, RPL30, RPL34, RPL37A, RPS6, RPL15, RPS13, RACK1, RPL41, RPL8, RPL6, RPS5, RPL37, RPS16, RPL11 |
199 |
| GSE42088_UNINF_VS_LEISHMANIA_INF_DC_2H_DN | 9.38e-36 | 107.26 | 57.88 | 7.62e-33 | 4.57e-32 | 24RPL13, RPL10, RPL32, RPS18, RPS15, RPS24, RPL36, RPL35A, RPL29, RPL7A, RPS7, RPS28, RPL24, RPL30, RPL19, RPS15A, RPS13, RACK1, RPL8, RPL6, RPL14, RPL37, RPS16, RPL11 |
200 |
| GSE14000_TRANSLATED_RNA_VS_MRNA_DC_DN | 4.59e-32 | 93.69 | 49.99 | 3.20e-29 | 2.24e-28 | 22RPL10, RPL32, RPS18, RPS3, RPL36, RPS14, RPLP1, RPL18, RPL30, RPS12, RPLP0, RPL13A, RPL19, RPSA, RPS15A, RPS6, RPS13, RACK1, RPL8, RPL12, RPS5, RPL11 |
194 |
| GSE42088_UNINF_VS_LEISHMANIA_INF_DC_4H_DN | 8.21e-32 | 91.05 | 48.63 | 5.00e-29 | 4.00e-28 | 22RPL13, RPL10, RPL32, RPS18, RPS19, RPS24, RPS14, RPL35A, RPLP1, RPS4X, RPL18, RPS7, RPL30, RPL34, RPL13A, RPS15A, RPS13, RACK1, RPL8, RPL37, RPS16, RPL11 |
199 |
| GSE22886_NAIVE_TCELL_VS_DC_UP | 7.20e-30 | 83.45 | 44.36 | 3.90e-27 | 3.51e-26 | 21RPS18, RPS23, RPS15, RPS27, RPS3, RPL36, RPS14, RPL35A, RPL29, RPL3, RPL7A, RPS7, RPL30, RPL34, RPL13A, RPL19, RPS15A, RPL6, RPL14, RPL11, RPL5 |
199 |
| GSE3720_UNSTIM_VS_PMA_STIM_VD2_GAMMADELTA_TCELL_UP | 3.47e-27 | 80.58 | 42.09 | 1.69e-24 | 1.69e-23 | 19RPS2, RPL32, RPS27, RPS3, RPL36, RPLP1, RPL18, RPL29, RPL3, RPL24, RPL30, RPS12, RPLP0, RPL37A, RPL19, RPSA, RPL12, RPL37, EEF1A1 |
175 |
| GSE3720_UNSTIM_VS_LPS_STIM_VD2_GAMMADELTA_TCELL_UP | 1.13e-25 | 77.43 | 39.97 | 5.01e-23 | 5.51e-22 | 18RPS2, RPL32, RPS19, RPS3, RPL36, RPL18, RPL29, RPL3, RPL30, RPS12, RPLP0, RPL37A, RPSA, RPL8, RPL12, RPL14, RPL37, RPL5 |
167 |
| GSE34205_HEALTHY_VS_FLU_INF_INFANT_PBMC_UP | 2.63e-24 | 64.12 | 33.24 | 1.07e-21 | 1.28e-20 | 18RPL13, RPL10, RPL32, RPS18, RPS23, RPS3A, RPS3, RPS14, RPLP1, RPL18, RPL7A, RPS12, RPL19, RPL15, RACK1, RPL8, RPS5, RPL5 |
198 |
| GSE3565_DUSP1_VS_WT_SPLENOCYTES_POST_LPS_INJECTION_UP | 2.71e-20 | 59.08 | 29.28 | 8.79e-18 | 1.32e-16 | 15RPL13, RPS2, RPL32, RPS15, RPL36, RPL35A, RPLP1, RPL18, RPL29, RPL24, RPLP0, RPL19, RPS15A, RPL8, RPS5 |
164 |
| GSE42724_NAIVE_VS_B1_BCELL_DN | 1.00e-20 | 52.75 | 26.70 | 3.75e-18 | 4.88e-17 | 16RPL13, RPS2, RPS3, RPL29, RPL3, RPL7A, RPS7, RPL34, RPL13A, RPSA, RPS6, RPL15, RACK1, RPL8, RPS5, RPL5 |
199 |
| GSE21927_SPLEEN_C57BL6_VS_4T1_TUMOR_BALBC_MONOCYTES_UP | 1.09e-20 | 52.46 | 26.56 | 3.78e-18 | 5.29e-17 | 16RPL13, RPS3A, RPS3, RPS27A, RPL35A, RPL18, RPL29, RPL3, RPL7A, RPS7, RPS15A, RPL15, RPL6, RPS5, RPL37, RPL5 |
200 |
| GSE26156_DOUBLE_POSITIVE_VS_CD4_SINGLE_POSITIVE_THYMOCYTE_UP | 2.38e-17 | 43.13 | 21.13 | 7.23e-15 | 1.16e-13 | 14RPL13, RPL10, RPL32, RPS23, RPS27A, RPL29, RPS28, RPL30, RPS12, RPL34, RPS15A, RPS13, RPL37, RPL11 |
199 |
| GSE24574_BCL6_HIGH_TFH_VS_TFH_CD4_TCELL_DN | 2.55e-17 | 42.89 | 21.02 | 7.30e-15 | 1.24e-13 | 14RPL13, RPL10, RPL32, RPS23, RPS27A, RPL29, RPS28, RPL30, RPS12, RPL34, RPS15A, RPS13, RPL37, RPS16 |
200 |
| GSE10239_NAIVE_VS_DAY4.5_EFF_CD8_TCELL_UP | 9.95e-16 | 38.77 | 18.60 | 2.69e-13 | 4.85e-12 | 13RPL13, RPL32, RPS18, RPS3, RPL36, RPS14, RPLP1, RPS7, RPLP0, RPL37A, RPL19, RPS15A, RPL11 |
199 |
| GSE5099_CLASSICAL_M1_VS_ALTERNATIVE_M2_MACROPHAGE_UP | 2.77e-14 | 35.61 | 16.66 | 7.11e-12 | 1.35e-10 | 12RPL13, RPL32, RPS18, RPS19, RPLP1, RPL29, RPL7A, RPL30, RPL13A, RPS15A, RPL12, RPL6 |
194 |
| GSE26488_WT_VS_VP16_TRANSGENIC_HDAC7_KO_DOUBLE_POSITIVE_THYMOCYTE_DN | 2.13e-13 | 36.63 | 16.61 | 4.95e-11 | 1.04e-09 | 11RPL13, RPL32, RPS14, RPS4X, RPL24, RPL37A, RPL19, RPS13, RPL12, RPS5, RPL37 |
169 |
Top Ranked Transcription Factors for this Gene Expression Program:
| Gene Symbol | Gene Loading Rank | DNA Binding Domain | Motif Status | IUPAC PWM | GTEx | DepMap | Decartes |
|---|---|---|---|---|---|---|---|
| RPS27A | 16 | No | ssDNA/RNA binding | Not a DNA binding protein | No motif | None | None |
| RPL7A | 23 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| RPL7 | 63 | No | Unlikely to be sequence specific TF | Low specificity DNA-binding protein | No motif | None | Binds DNA by filter binding (PMID: 8441630), but competition assays suggest it is non-specific. A specific dsRNA structure is thought to be the preferred substrate. RNA-binding has been demonstrated by gel-shift experiments (PMID: 10329420). |
| UBA52 | 76 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| NPM1 | 90 | No | Unlikely to be sequence specific TF | Low specificity DNA-binding protein | No motif | None | No evidence for sequence-specific DNA-binding (PMID: 2223875) |
| YBX1 | 92 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | Might also bind RNA |
| HNRNPA1 | 93 | No | ssDNA/RNA binding | Not a DNA binding protein | No motif | None | Binds single stranded DNA in the structure (PDB: 1PGZ) |
| RPL36A | 107 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Protein is a ribosomal component |
| PHB2 | 117 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | Protein has been shown to bind to TFs and function as a co-repressor. There appears to be no evidence for direct DNA-binding activity (PMID: 18629613) |
| COMMD6 | 139 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| HMGB1 | 142 | No | Unlikely to be sequence specific TF | Low specificity DNA-binding protein | No motif | None | In the alignment, it clusters with other HMG proteins that have not yielded motifs. Evidence exists showing HMGB½ are non-sequence-specific binding proteins (PMID: 11497996). |
| SOX11 | 146 | Yes | Known motif | Monomer or homomultimer | 100 perc ID - in vitro | None | None |
| ZNF428 | 158 | Yes | Likely to be sequence specific TF | Monomer or homomultimer | No motif | Single C2H2 domain | None |
| PHOX2B | 160 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| TWIST1 | 161 | Yes | Known motif | Monomer or homomultimer | In vivo/Misc source | Only known motifs are from Transfac or HocoMoco - origin is uncertain | Can form both homodimers and heterodimers with TCF3 (PMID: 16502419). |
| MYCN | 185 | Yes | Known motif | Monomer or homomultimer | High-throughput in vitro | None | None |
| EIF3K | 191 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| UBB | 216 | No | Unlikely to be sequence specific TF | Not a DNA binding protein | No motif | None | None |
| HMGA1 | 222 | Yes | Known motif | Monomer or homomultimer | In vivo/Misc source | Only known motifs are from Transfac or HocoMoco - origin is uncertain | None |
| PCBP2 | 226 | No | ssDNA/RNA binding | Not a DNA binding protein | No motif | None | Protein is an RBP. Structure (PDB:2PQU) is with single stranded DNA |
Druggable Genes for this Gene Expression Program:
| Gene Symbol | Gene Loading Rank | Drug Names | Drug Category | GTEx | DepMap |
|---|---|---|---|---|---|
| TUBB2B | 87 | 9VINCRISTINE, VINORELBINE BASE, ERIBULIN, CABAZITAXEL, IXABEPILONE, DOCETAXEL, COLCHICINE, PACLITAXEL, VINBLASTINE |
Small molecule | GTEx | DepMap |
| TUBB2B | 87 | 2BRENTUXIMAB VEDOTIN, TRASTUZUMAB EMTANSINE |
Antibody | GTEx | DepMap |
| TUBB | 99 | 9CABAZITAXEL, VINORELBINE BASE, VINCRISTINE, ERIBULIN, IXABEPILONE, DOCETAXEL, COLCHICINE, PACLITAXEL, VINBLASTINE |
Small molecule | GTEx | DepMap |
| TUBB | 99 | 2BRENTUXIMAB VEDOTIN, TRASTUZUMAB EMTANSINE |
Antibody | GTEx | DepMap |
| IMPDH2 | 123 | 3MYCOPHENOLATE MOFETIL, MYCOPHENOLIC ACID, THIOGUANINE |
Small molecule | GTEx | DepMap |
| NDUFB11 | 127 | 1METFORMIN |
Small molecule | GTEx | DepMap |
| NDUFB7 | 154 | 1METFORMIN |
Small molecule | GTEx | DepMap |
| NDUFC2 | 204 | 1METFORMIN |
Small molecule | GTEx | DepMap |
| NDUFS5 | 251 | 1METFORMIN |
Small molecule | GTEx | DepMap |
| NDUFB10 | 256 | 1METFORMIN |
Small molecule | GTEx | DepMap |
| ODC1 | 263 | 1EFLORNITHINE |
Small molecule | GTEx | DepMap |
| HDAC2 | 276 | 4ROMIDEPSIN, PANOBINOSTAT, VORINOSTAT, BELINOSTAT |
Small molecule | GTEx | DepMap |
| NDUFB4 | 296 | 1METFORMIN |
Small molecule | GTEx | DepMap |
| NDUFB2 | 300 | 1METFORMIN |
Small molecule | GTEx | DepMap |
| NDUFB8 | 311 | 1METFORMIN |
Small molecule | GTEx | DepMap |
| NDUFA11 | 355 | 1METFORMIN |
Small molecule | GTEx | DepMap |
| NDUFS6 | 363 | 1METFORMIN |
Small molecule | GTEx | DepMap |
| MAP2K2 | 374 | 2COBIMETINIB, TRAMETINIB |
Small molecule | GTEx | DepMap |
| NDUFB1 | 391 | 1METFORMIN |
Small molecule | GTEx | DepMap |
| BSG | 392 | 1GAVILIMOMAB |
Antibody | GTEx | DepMap |
| FASN | 452 | 1ORLISTAT |
Small molecule | GTEx | DepMap |
| NDUFA2 | 459 | 1METFORMIN |
Small molecule | GTEx | DepMap |
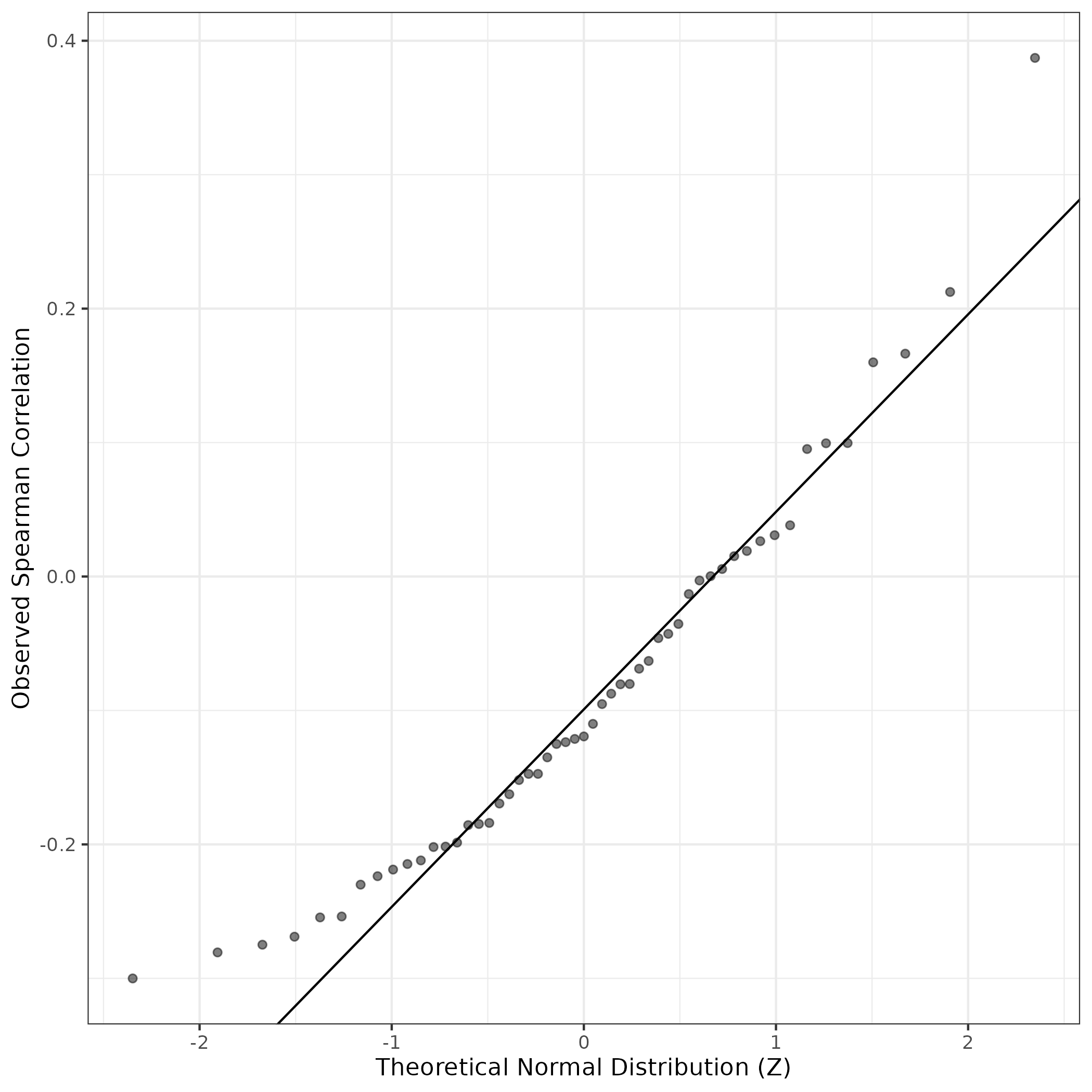
QQ Plot showing correlations with other GEPs in this dataset, calculated by Spearman correlation:
Interactive QQ-plot of gene loadings:
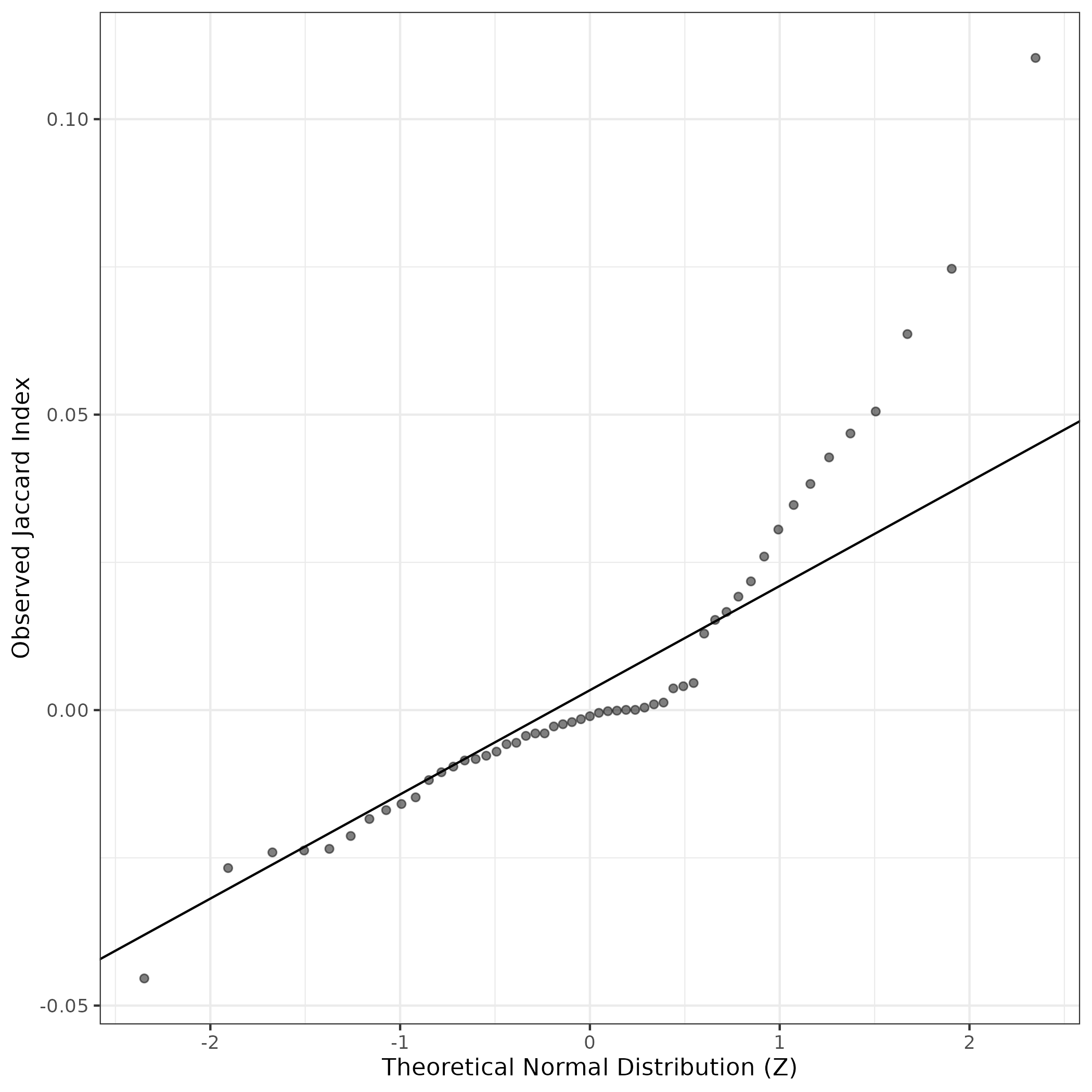
A similar QQ-plot as above, but only for instances where the H value is e.g. > 25, i.e. we are confident that the expression program is active above noise. Agreemenet between these binary vectors is tested using the Jaccard Index, with the P-values calculated by an exact test:
Interactive QQ-plot:
Singler cell type annotations for the top 50 cells on this program.
| Cell ID | Singler label | Singler Delta | Activity Score | Top Singler Raw Scores |
|---|---|---|---|---|
| TACCTCGTCTGCTAGA-1_HTA4_1021_4082 | Neurons | 0.28 | 1828.95 | Raw ScoresNeurons: 0.53, Neuroepithelial_cell: 0.47, Astrocyte: 0.47, Embryonic_stem_cells: 0.42, iPS_cells: 0.41, MSC: 0.36, Smooth_muscle_cells: 0.34, Fibroblasts: 0.34, Tissue_stem_cells: 0.33, Endothelial_cells: 0.33 |
| GTTGTAGAGATGGCAC-1_HTA4_1021_4082 | Neurons | 0.27 | 777.92 | Raw ScoresNeurons: 0.48, Neuroepithelial_cell: 0.44, Astrocyte: 0.41, Embryonic_stem_cells: 0.37, iPS_cells: 0.36, MSC: 0.31, Tissue_stem_cells: 0.28, Fibroblasts: 0.27, Endothelial_cells: 0.27, Smooth_muscle_cells: 0.26 |
| GGAGAACGTGACAACG-1_HTA4_1021_4082 | Neurons | 0.29 | 758.19 | Raw ScoresNeurons: 0.52, Neuroepithelial_cell: 0.47, Astrocyte: 0.46, Embryonic_stem_cells: 0.42, iPS_cells: 0.41, MSC: 0.36, Tissue_stem_cells: 0.32, Fibroblasts: 0.32, Endothelial_cells: 0.31, Smooth_muscle_cells: 0.31 |
| TCCTCGAGTCAAGTTC-1_HTA4_1021_4082 | Neurons | 0.24 | 753.16 | Raw ScoresNeurons: 0.43, Neuroepithelial_cell: 0.38, Astrocyte: 0.36, Embryonic_stem_cells: 0.34, iPS_cells: 0.34, MSC: 0.27, Fibroblasts: 0.26, Smooth_muscle_cells: 0.25, Endothelial_cells: 0.25, Osteoblasts: 0.25 |
| GGTTGTACAACCGTAT-1_HTA4_1021_4082 | Neurons | 0.29 | 738.99 | Raw ScoresNeurons: 0.53, Astrocyte: 0.46, Neuroepithelial_cell: 0.46, Embryonic_stem_cells: 0.41, iPS_cells: 0.4, MSC: 0.36, Smooth_muscle_cells: 0.33, Tissue_stem_cells: 0.33, Endothelial_cells: 0.32, Fibroblasts: 0.32 |
| ACGGAAGCATCTGCGG-1_HTA4_1021_4082 | Neuroepithelial_cell | 0.11 | 683.27 | Raw ScoresNeurons: 0.3, Neuroepithelial_cell: 0.29, Astrocyte: 0.28, Embryonic_stem_cells: 0.26, iPS_cells: 0.25, Smooth_muscle_cells: 0.23, MSC: 0.23, Fibroblasts: 0.22, Tissue_stem_cells: 0.22, Osteoblasts: 0.22 |
| ATGGGTTAGTCCCAGC-1_HTA4_1021_4082 | Neurons | 0.20 | 680.01 | Raw ScoresNeurons: 0.36, Neuroepithelial_cell: 0.32, Astrocyte: 0.31, Embryonic_stem_cells: 0.27, iPS_cells: 0.26, MSC: 0.21, Endothelial_cells: 0.2, Smooth_muscle_cells: 0.2, Fibroblasts: 0.2, Tissue_stem_cells: 0.19 |
| ATTGGGTAGAGAGTTT-1_HTA4_1008_4030 | Neurons | 0.27 | 624.11 | Raw ScoresNeurons: 0.5, Neuroepithelial_cell: 0.46, Astrocyte: 0.44, Embryonic_stem_cells: 0.43, iPS_cells: 0.41, MSC: 0.36, Tissue_stem_cells: 0.31, Endothelial_cells: 0.3, Fibroblasts: 0.3, Smooth_muscle_cells: 0.3 |
| TGTAACGGTGATAGAT-1_HTA4_1021_4082 | Neurons | 0.19 | 617.38 | Raw ScoresNeurons: 0.33, Neuroepithelial_cell: 0.28, Astrocyte: 0.28, Embryonic_stem_cells: 0.25, iPS_cells: 0.24, MSC: 0.2, Smooth_muscle_cells: 0.2, Osteoblasts: 0.2, Chondrocytes: 0.2, Fibroblasts: 0.2 |
| TCGCACTTCCTTGACC-1_HTA4_1021_4083 | Neurons | 0.13 | 569.32 | Raw ScoresEndothelial_cells: 0.37, Neuroepithelial_cell: 0.35, Astrocyte: 0.34, Neurons: 0.34, MSC: 0.33, Fibroblasts: 0.33, Smooth_muscle_cells: 0.32, Osteoblasts: 0.32, Embryonic_stem_cells: 0.32, Tissue_stem_cells: 0.31 |
| CTTCTCTTCTTCACAT-1_HTA4_1021_4083 | Neurons | 0.18 | 561.42 | Raw ScoresNeurons: 0.3, Neuroepithelial_cell: 0.27, Astrocyte: 0.26, Embryonic_stem_cells: 0.24, iPS_cells: 0.23, MSC: 0.21, Tissue_stem_cells: 0.18, Endothelial_cells: 0.18, Fibroblasts: 0.18, Smooth_muscle_cells: 0.17 |
| TCCCACACACGGTGTC-1_HTA4_1005_4018 | Neurons | 0.26 | 524.24 | Raw ScoresNeurons: 0.39, Astrocyte: 0.35, Neuroepithelial_cell: 0.34, Embryonic_stem_cells: 0.29, iPS_cells: 0.28, MSC: 0.23, Smooth_muscle_cells: 0.22, Osteoblasts: 0.21, Fibroblasts: 0.21, Chondrocytes: 0.2 |
| CCGATGGGTCGAGCTC-1_HTA4_1021_4083 | Neurons | 0.21 | 488.19 | Raw ScoresNeurons: 0.38, Neuroepithelial_cell: 0.33, Astrocyte: 0.32, Embryonic_stem_cells: 0.29, iPS_cells: 0.28, MSC: 0.25, Fibroblasts: 0.22, Endothelial_cells: 0.22, Smooth_muscle_cells: 0.22, Tissue_stem_cells: 0.21 |
| GGTGTTATCGTTGTAG-1_HTA4_1021_4082 | Neurons | 0.21 | 470.71 | Raw ScoresNeurons: 0.4, Neuroepithelial_cell: 0.34, Astrocyte: 0.33, iPS_cells: 0.3, Embryonic_stem_cells: 0.3, MSC: 0.24, Fibroblasts: 0.23, Smooth_muscle_cells: 0.23, Endothelial_cells: 0.23, Osteoblasts: 0.23 |
| TACGCTCAGAGGCGTT-1_HTA4_1021_4082 | Neurons | 0.20 | 465.81 | Raw ScoresNeurons: 0.39, Neuroepithelial_cell: 0.35, Astrocyte: 0.33, Embryonic_stem_cells: 0.31, iPS_cells: 0.3, MSC: 0.25, Endothelial_cells: 0.22, Tissue_stem_cells: 0.22, Fibroblasts: 0.21, Pro-B_cell_CD34+: 0.21 |
| TCTATCAAGAAACTAC-1_HTA4_1001_4004 | Smooth_muscle_cells | 0.15 | 465.58 | Raw ScoresFibroblasts: 0.44, Smooth_muscle_cells: 0.44, Osteoblasts: 0.42, Tissue_stem_cells: 0.42, MSC: 0.42, Chondrocytes: 0.41, Endothelial_cells: 0.41, iPS_cells: 0.41, Astrocyte: 0.4, Neurons: 0.4 |
| GATTCTTCAGAAGTGC-1_HTA4_1021_4082 | Neurons | 0.21 | 451.99 | Raw ScoresNeurons: 0.39, Neuroepithelial_cell: 0.35, Astrocyte: 0.33, Embryonic_stem_cells: 0.3, iPS_cells: 0.3, MSC: 0.25, Endothelial_cells: 0.24, Smooth_muscle_cells: 0.23, Tissue_stem_cells: 0.23, Fibroblasts: 0.22 |
| TCACGGGCATACTGAC-1_HTA4_1021_4082 | Neurons | 0.11 | 449.01 | Raw ScoresNeurons: 0.23, Neuroepithelial_cell: 0.21, iPS_cells: 0.2, Embryonic_stem_cells: 0.2, Astrocyte: 0.2, MSC: 0.17, Endothelial_cells: 0.15, Smooth_muscle_cells: 0.15, Tissue_stem_cells: 0.15, Fibroblasts: 0.14 |
| GAGAGGTCAACTTGCA-1_HTA4_1021_4082 | Neurons | 0.15 | 448.57 | Raw ScoresNeurons: 0.29, Neuroepithelial_cell: 0.27, Astrocyte: 0.26, Embryonic_stem_cells: 0.24, iPS_cells: 0.22, MSC: 0.2, Endothelial_cells: 0.19, Fibroblasts: 0.19, Smooth_muscle_cells: 0.18, Chondrocytes: 0.18 |
| CTACAGAAGGTACAAT-1_HTA4_1021_4083 | Neurons | 0.13 | 441.79 | Raw ScoresNeurons: 0.3, Neuroepithelial_cell: 0.27, Embryonic_stem_cells: 0.25, iPS_cells: 0.24, Astrocyte: 0.24, MSC: 0.21, Endothelial_cells: 0.19, Fibroblasts: 0.19, Pro-B_cell_CD34+: 0.19, CMP: 0.18 |
| GAGGGTAGTCTAGGTT-1_HTA4_1021_4082 | Neurons | 0.18 | 437.57 | Raw ScoresNeurons: 0.32, Neuroepithelial_cell: 0.27, Astrocyte: 0.26, Embryonic_stem_cells: 0.24, iPS_cells: 0.23, MSC: 0.21, Endothelial_cells: 0.19, Smooth_muscle_cells: 0.19, Fibroblasts: 0.19, Tissue_stem_cells: 0.19 |
| CTTCTAACAAACTCTG-1_HTA4_1008_4030 | Neurons | 0.28 | 434.67 | Raw ScoresNeurons: 0.5, Neuroepithelial_cell: 0.45, Astrocyte: 0.43, Embryonic_stem_cells: 0.4, iPS_cells: 0.38, MSC: 0.35, Tissue_stem_cells: 0.31, Fibroblasts: 0.3, Smooth_muscle_cells: 0.3, Endothelial_cells: 0.3 |
| ATTCTTGTCACAGAGG-1_HTA4_1001_4004 | Smooth_muscle_cells | 0.14 | 431.53 | Raw ScoresFibroblasts: 0.44, Smooth_muscle_cells: 0.43, Tissue_stem_cells: 0.42, MSC: 0.42, Osteoblasts: 0.42, Astrocyte: 0.41, iPS_cells: 0.41, Chondrocytes: 0.41, Neurons: 0.41, Endothelial_cells: 0.4 |
| GTTACGACAAAGTGTA-1_HTA4_1021_4082 | Neurons | 0.13 | 417.79 | Raw ScoresNeurons: 0.22, Astrocyte: 0.21, Neuroepithelial_cell: 0.2, Embryonic_stem_cells: 0.16, iPS_cells: 0.16, Smooth_muscle_cells: 0.13, MSC: 0.13, Endothelial_cells: 0.12, Chondrocytes: 0.11, Tissue_stem_cells: 0.11 |
| AACTTCTTCTGTGTGA-1_HTA4_1021_4082 | Neurons | 0.22 | 416.78 | Raw ScoresNeurons: 0.42, Neuroepithelial_cell: 0.36, Astrocyte: 0.34, Embryonic_stem_cells: 0.32, iPS_cells: 0.32, MSC: 0.27, Endothelial_cells: 0.26, Smooth_muscle_cells: 0.25, Fibroblasts: 0.25, Tissue_stem_cells: 0.24 |
| CCAATGAAGCGCCGTT-1_HTA4_1021_4083 | Neurons | 0.25 | 416.29 | Raw ScoresNeurons: 0.43, Neuroepithelial_cell: 0.38, Astrocyte: 0.37, Embryonic_stem_cells: 0.34, iPS_cells: 0.33, MSC: 0.28, Smooth_muscle_cells: 0.25, Endothelial_cells: 0.25, Fibroblasts: 0.25, Tissue_stem_cells: 0.25 |
| TTACCGCAGATAGTCA-1_HTA4_1021_4082 | Neurons | 0.27 | 415.46 | Raw ScoresNeurons: 0.48, Astrocyte: 0.44, Neuroepithelial_cell: 0.43, Embryonic_stem_cells: 0.37, iPS_cells: 0.35, MSC: 0.33, Fibroblasts: 0.32, Smooth_muscle_cells: 0.32, Endothelial_cells: 0.31, Osteoblasts: 0.31 |
| AGTCACAAGACCAAAT-1_HTA4_1005_4018 | Neurons | 0.23 | 413.87 | Raw ScoresNeurons: 0.36, Neuroepithelial_cell: 0.35, Astrocyte: 0.33, Embryonic_stem_cells: 0.29, iPS_cells: 0.28, MSC: 0.23, Smooth_muscle_cells: 0.22, Osteoblasts: 0.22, Fibroblasts: 0.21, Chondrocytes: 0.2 |
| TGGTAGTCAACTCCAA-1_HTA4_1021_4082 | Neurons | 0.23 | 412.40 | Raw ScoresNeurons: 0.42, Neuroepithelial_cell: 0.37, Astrocyte: 0.37, Embryonic_stem_cells: 0.32, iPS_cells: 0.31, MSC: 0.27, Endothelial_cells: 0.25, Smooth_muscle_cells: 0.24, Tissue_stem_cells: 0.23, Fibroblasts: 0.23 |
| TCCCATGAGCGGTAAC-1_HTA4_1010_4035 | Neurons | 0.21 | 409.32 | Raw ScoresNeurons: 0.34, Neuroepithelial_cell: 0.29, Astrocyte: 0.28, Embryonic_stem_cells: 0.25, iPS_cells: 0.23, Smooth_muscle_cells: 0.21, MSC: 0.21, Endothelial_cells: 0.2, Fibroblasts: 0.2, Osteoblasts: 0.19 |
| AGTCATGCATTCGGGC-1_HTA4_1021_4082 | Neurons | 0.17 | 401.65 | Raw ScoresNeurons: 0.3, Neuroepithelial_cell: 0.27, Astrocyte: 0.24, Embryonic_stem_cells: 0.24, iPS_cells: 0.23, MSC: 0.19, Fibroblasts: 0.17, Tissue_stem_cells: 0.16, Smooth_muscle_cells: 0.16, Endothelial_cells: 0.15 |
| GTCTTTACAGATTCGT-1_HTA4_1021_4082 | Neurons | 0.21 | 397.34 | Raw ScoresNeurons: 0.36, Neuroepithelial_cell: 0.32, Astrocyte: 0.3, Embryonic_stem_cells: 0.27, iPS_cells: 0.26, MSC: 0.23, Fibroblasts: 0.22, Osteoblasts: 0.21, Smooth_muscle_cells: 0.21, Tissue_stem_cells: 0.2 |
| TCTAACTGTTGTCAGT-1_HTA4_1021_4082 | Neurons | 0.19 | 394.39 | Raw ScoresNeurons: 0.33, Neuroepithelial_cell: 0.28, Astrocyte: 0.27, Embryonic_stem_cells: 0.25, iPS_cells: 0.23, MSC: 0.2, Smooth_muscle_cells: 0.19, Endothelial_cells: 0.18, Tissue_stem_cells: 0.18, Fibroblasts: 0.17 |
| TTCCGGTGTCATAAAG-1_HTA4_1021_4082 | Neurons | 0.21 | 394.35 | Raw ScoresNeurons: 0.41, Neuroepithelial_cell: 0.36, Astrocyte: 0.35, Embryonic_stem_cells: 0.33, iPS_cells: 0.32, MSC: 0.27, Fibroblasts: 0.26, Smooth_muscle_cells: 0.26, Endothelial_cells: 0.26, Tissue_stem_cells: 0.25 |
| ATACCTTTCCCTATTA-1_HTA4_1021_4082 | Neurons | 0.21 | 378.31 | Raw ScoresNeurons: 0.36, Neuroepithelial_cell: 0.32, Astrocyte: 0.29, Embryonic_stem_cells: 0.27, iPS_cells: 0.27, MSC: 0.22, Smooth_muscle_cells: 0.19, Fibroblasts: 0.19, Endothelial_cells: 0.19, Tissue_stem_cells: 0.19 |
| AGAGCAGGTACTAACC-1_HTA4_1021_4083 | Neurons | 0.23 | 376.12 | Raw ScoresNeurons: 0.4, Neuroepithelial_cell: 0.35, Astrocyte: 0.34, Embryonic_stem_cells: 0.31, iPS_cells: 0.3, MSC: 0.25, Endothelial_cells: 0.23, Tissue_stem_cells: 0.23, Fibroblasts: 0.23, Smooth_muscle_cells: 0.22 |
| ACGGAAGGTTACGGAG-1_HTA4_1008_4027 | Neurons | 0.27 | 370.48 | Raw ScoresNeurons: 0.47, Neuroepithelial_cell: 0.41, Astrocyte: 0.4, Embryonic_stem_cells: 0.37, iPS_cells: 0.36, MSC: 0.31, Smooth_muscle_cells: 0.28, Tissue_stem_cells: 0.27, Fibroblasts: 0.27, Endothelial_cells: 0.27 |
| GATGTTGTCCTCTCGA-1_HTA4_1021_4083 | Neurons | 0.16 | 369.49 | Raw ScoresNeurons: 0.32, Neuroepithelial_cell: 0.31, Astrocyte: 0.28, Embryonic_stem_cells: 0.27, iPS_cells: 0.26, MSC: 0.22, Fibroblasts: 0.21, Endothelial_cells: 0.2, Smooth_muscle_cells: 0.2, Tissue_stem_cells: 0.2 |
| ATTCGTTCACGCCACA-1_HTA4_1021_4082 | Neurons | 0.24 | 368.09 | Raw ScoresNeurons: 0.44, Neuroepithelial_cell: 0.4, Astrocyte: 0.38, Embryonic_stem_cells: 0.34, iPS_cells: 0.33, MSC: 0.28, Endothelial_cells: 0.26, Tissue_stem_cells: 0.24, Fibroblasts: 0.24, Smooth_muscle_cells: 0.23 |
| TGTTCATCAATCAGCT-1_HTA4_1021_4082 | Neurons | 0.24 | 362.96 | Raw ScoresNeurons: 0.42, Neuroepithelial_cell: 0.38, Astrocyte: 0.37, Embryonic_stem_cells: 0.33, iPS_cells: 0.32, MSC: 0.27, Tissue_stem_cells: 0.24, Smooth_muscle_cells: 0.24, Endothelial_cells: 0.24, Fibroblasts: 0.23 |
| CGCAGGTGTAGCCCTG-1_HTA4_1021_4083 | Neurons | 0.24 | 352.52 | Raw ScoresNeurons: 0.43, Neuroepithelial_cell: 0.39, Astrocyte: 0.37, Embryonic_stem_cells: 0.34, iPS_cells: 0.33, MSC: 0.28, Fibroblasts: 0.26, Smooth_muscle_cells: 0.26, Tissue_stem_cells: 0.26, Osteoblasts: 0.25 |
| TCGCTCAGTGTTACTG-1_HTA4_1021_4082 | Neurons | 0.20 | 351.37 | Raw ScoresNeurons: 0.38, Neuroepithelial_cell: 0.34, Astrocyte: 0.31, Embryonic_stem_cells: 0.3, iPS_cells: 0.29, MSC: 0.25, Endothelial_cells: 0.22, Smooth_muscle_cells: 0.21, Tissue_stem_cells: 0.21, Fibroblasts: 0.21 |
| GGTGATTAGCCTCTGG-1_HTA4_1021_4082 | Neurons | 0.21 | 348.35 | Raw ScoresNeurons: 0.36, Neuroepithelial_cell: 0.33, Astrocyte: 0.32, Embryonic_stem_cells: 0.28, iPS_cells: 0.26, MSC: 0.24, Smooth_muscle_cells: 0.21, Tissue_stem_cells: 0.21, Fibroblasts: 0.21, Endothelial_cells: 0.2 |
| TTGAACGAGGTAGATT-1_HTA4_1021_4083 | Neurons | 0.18 | 345.50 | Raw ScoresNeurons: 0.34, Neuroepithelial_cell: 0.31, Astrocyte: 0.28, Embryonic_stem_cells: 0.26, iPS_cells: 0.25, MSC: 0.22, Endothelial_cells: 0.2, Smooth_muscle_cells: 0.2, Tissue_stem_cells: 0.19, Fibroblasts: 0.18 |
| TGCTTCGTCGGCGATC-1_HTA4_1021_4083 | Neurons | 0.22 | 343.61 | Raw ScoresNeurons: 0.41, Neuroepithelial_cell: 0.37, Astrocyte: 0.36, Embryonic_stem_cells: 0.33, iPS_cells: 0.32, MSC: 0.27, Endothelial_cells: 0.24, Smooth_muscle_cells: 0.24, Fibroblasts: 0.24, Tissue_stem_cells: 0.24 |
| GTGCGTGCAACTACGT-1_HTA4_1010_4035 | Neurons | 0.20 | 335.34 | Raw ScoresNeurons: 0.31, Neuroepithelial_cell: 0.28, Astrocyte: 0.27, Embryonic_stem_cells: 0.24, iPS_cells: 0.23, MSC: 0.2, Smooth_muscle_cells: 0.18, Fibroblasts: 0.18, Osteoblasts: 0.17, Endothelial_cells: 0.17 |
| CAGCGTGCAGACATCT-1_HTA4_1008_4028 | Neurons | 0.29 | 335.33 | Raw ScoresNeurons: 0.51, Neuroepithelial_cell: 0.47, Astrocyte: 0.45, Embryonic_stem_cells: 0.42, iPS_cells: 0.4, MSC: 0.36, Tissue_stem_cells: 0.32, Endothelial_cells: 0.3, Fibroblasts: 0.3, Smooth_muscle_cells: 0.3 |
| GTCATTTAGCATATGA-1_HTA4_1021_4082 | Neurons | 0.16 | 332.96 | Raw ScoresNeurons: 0.28, Neuroepithelial_cell: 0.25, Astrocyte: 0.23, Embryonic_stem_cells: 0.22, iPS_cells: 0.21, MSC: 0.18, Endothelial_cells: 0.18, Fibroblasts: 0.17, Smooth_muscle_cells: 0.17, Osteoblasts: 0.17 |
| GGGACAATCAGGTAAA-1_HTA4_1021_4082 | Neurons | 0.25 | 323.89 | Raw ScoresNeurons: 0.41, Neuroepithelial_cell: 0.37, Astrocyte: 0.35, Embryonic_stem_cells: 0.32, iPS_cells: 0.3, MSC: 0.26, Endothelial_cells: 0.24, Smooth_muscle_cells: 0.24, Fibroblasts: 0.23, Tissue_stem_cells: 0.23 |
| TGCTTGCAGCGAGTCA-1_HTA4_1021_4082 | Neurons | 0.24 | 320.46 | Raw ScoresNeurons: 0.46, Neuroepithelial_cell: 0.4, Astrocyte: 0.39, Embryonic_stem_cells: 0.35, iPS_cells: 0.34, MSC: 0.29, Endothelial_cells: 0.27, Fibroblasts: 0.27, Tissue_stem_cells: 0.27, Smooth_muscle_cells: 0.26 |
Below shows the significant enrichments of this GEP for literature curated gene lists
This data was procured from existing single cell RNA-seq maps of neuroblastoma or related relevant data.
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by the P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Proliferating Cells
These are canonical marker genes of cycling cells, obtained from Fig. 3 of Hsiao et al. (PMID 32312741):
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.05e-05
Mean rank of genes in gene set: 1210.67
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| H4C3 | 0.0058577 | 89 | GTEx | DepMap | Descartes | 0.66 | NA |
| UBE2C | 0.0013856 | 688 | GTEx | DepMap | Descartes | 0.25 | 190.35 |
| TOP2A | 0.0010713 | 989 | GTEx | DepMap | Descartes | 0.79 | 88.56 |
| MKI67 | 0.0006942 | 1645 | GTEx | DepMap | Descartes | 0.28 | 14.02 |
| H4C5 | 0.0006049 | 1882 | GTEx | DepMap | Descartes | 0.03 | NA |
| CDK1 | 0.0005713 | 1971 | GTEx | DepMap | Descartes | 0.18 | 59.72 |
Neuroblast (Hanemaaijer)
Marker genes obtained from Supplementary Table SD of Hanemaaijer et al (PMID 33500353). The authors generated single-cell RNA-seq data (sort-seq, 2,229 cells total) from mouse adrenal glads at E13.5, E14.5, E17.5, E18.5, P1 and P5. These were marker genes that matched with a similar dataset generated by Furlan et al (PMID 28684471). This particular set of markers are for the Neuroblast subcluster, which is part of the Adrenal Medulla cluster.:
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.16e-04
Mean rank of genes in gene set: 4228.92
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| STMN1 | 0.0044213 | 114 | GTEx | DepMap | Descartes | 2.61 | 535.43 |
| STMN2 | 0.0033906 | 159 | GTEx | DepMap | Descartes | 8.67 | 2137.58 |
| BASP1 | 0.0033407 | 166 | GTEx | DepMap | Descartes | 2.71 | 809.68 |
| CCND1 | 0.0032207 | 181 | GTEx | DepMap | Descartes | 2.62 | 340.49 |
| ISL1 | 0.0021995 | 339 | GTEx | DepMap | Descartes | 0.94 | 221.35 |
| INA | 0.0018520 | 450 | GTEx | DepMap | Descartes | 0.55 | 89.12 |
| ELAVL4 | 0.0016600 | 527 | GTEx | DepMap | Descartes | 3.08 | 368.17 |
| ELAVL3 | 0.0010804 | 978 | GTEx | DepMap | Descartes | 1.41 | 148.04 |
| RTN1 | 0.0009450 | 1162 | GTEx | DepMap | Descartes | 2.42 | 338.82 |
| TUBB3 | 0.0001827 | 4050 | GTEx | DepMap | Descartes | 0.03 | 6.29 |
| NEFM | 0.0000023 | 6470 | GTEx | DepMap | Descartes | 0.84 | 104.26 |
| NEFL | -0.0013195 | 19903 | GTEx | DepMap | Descartes | 1.19 | 103.59 |
| PRPH | -0.0019761 | 20477 | GTEx | DepMap | Descartes | 1.52 | 247.51 |
Adrenal Premordium (Hanemaaijer)
Marker genes obtained from Supplementary Table SD of Hanemaaijer et al (PMID 33500353). The authors generated single-cell RNA-seq data (sort-seq, 2,229 cells total) from mouse adrenal glads at E13.5, E14.5, E17.5, E18.5, P1 and P5. These were marker genes that matched with a similar dataset generated by Furlan et al (PMID 28684471). This particular set of markers are for the Adrenal Premordium subcluster, which is part of the Cortex cluster.:
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.23e-04
Mean rank of genes in gene set: 548.6
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| RPS2 | 0.0160635 | 3 | GTEx | DepMap | Descartes | 10.51 | 4193.09 |
| NPM1 | 0.0058507 | 90 | GTEx | DepMap | Descartes | 1.91 | 626.54 |
| MIF | 0.0028128 | 225 | GTEx | DepMap | Descartes | 0.46 | 265.30 |
| TPI1 | 0.0027771 | 229 | GTEx | DepMap | Descartes | 0.95 | 294.00 |
| TK1 | 0.0005089 | 2196 | GTEx | DepMap | Descartes | 0.09 | 32.69 |
Below shows ranks on this GEP for literature curated gene lists for large gene sets
These include those reported as mesenchymal/adrenergic by Van Groningen et al.
High ranks indicate this gene is a driver of this GEP (note these results are not ordered).
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
VanGroningen Adrenergic Genes
Adrenergic marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.42e-18
Mean rank of genes in gene set: 7578.86
Median rank of genes in gene set: 3750
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TUBB2B | 0.0060284 | 87 | GTEx | DepMap | Descartes | 3.63 | 1054.34 |
| DLK1 | 0.0058787 | 88 | GTEx | DepMap | Descartes | 1.42 | 166.70 |
| EEF1A2 | 0.0051036 | 100 | GTEx | DepMap | Descartes | 2.05 | 270.68 |
| CCNI | 0.0040684 | 129 | GTEx | DepMap | Descartes | 2.32 | 468.11 |
| SOX11 | 0.0035339 | 146 | GTEx | DepMap | Descartes | 2.22 | 145.51 |
| CKB | 0.0035270 | 147 | GTEx | DepMap | Descartes | 1.20 | 446.28 |
| STMN2 | 0.0033906 | 159 | GTEx | DepMap | Descartes | 8.67 | 2137.58 |
| PHOX2B | 0.0033789 | 160 | GTEx | DepMap | Descartes | 1.63 | 332.46 |
| NPY | 0.0032694 | 171 | GTEx | DepMap | Descartes | 3.49 | 2692.04 |
| RANBP1 | 0.0032601 | 172 | GTEx | DepMap | Descartes | 0.74 | 211.20 |
| CCND1 | 0.0032207 | 181 | GTEx | DepMap | Descartes | 2.62 | 340.49 |
| EIF1B | 0.0032012 | 182 | GTEx | DepMap | Descartes | 0.72 | 360.93 |
| CHGA | 0.0029321 | 211 | GTEx | DepMap | Descartes | 1.08 | 286.09 |
| HNRNPA0 | 0.0028918 | 215 | GTEx | DepMap | Descartes | 0.73 | 45.96 |
| HMGA1 | 0.0028512 | 222 | GTEx | DepMap | Descartes | 0.42 | 122.03 |
| FABP6 | 0.0026956 | 245 | GTEx | DepMap | Descartes | 0.31 | 199.47 |
| GDAP1L1 | 0.0026847 | 247 | GTEx | DepMap | Descartes | 0.56 | 105.11 |
| LSM3 | 0.0026671 | 249 | GTEx | DepMap | Descartes | 0.40 | 64.74 |
| C4orf48 | 0.0026335 | 252 | GTEx | DepMap | Descartes | 0.94 | 1001.73 |
| PHPT1 | 0.0025992 | 255 | GTEx | DepMap | Descartes | 0.65 | 262.82 |
| GAL | 0.0025011 | 267 | GTEx | DepMap | Descartes | 2.19 | 1228.63 |
| RBP1 | 0.0023971 | 290 | GTEx | DepMap | Descartes | 0.53 | 137.11 |
| RGS5 | 0.0023335 | 307 | GTEx | DepMap | Descartes | 2.31 | 203.77 |
| DCX | 0.0022537 | 326 | GTEx | DepMap | Descartes | 0.77 | 45.02 |
| ISL1 | 0.0021995 | 339 | GTEx | DepMap | Descartes | 0.94 | 221.35 |
| LSM4 | 0.0021670 | 348 | GTEx | DepMap | Descartes | 0.64 | 207.05 |
| STMN4 | 0.0021356 | 357 | GTEx | DepMap | Descartes | 1.07 | 232.45 |
| CENPV | 0.0020648 | 381 | GTEx | DepMap | Descartes | 0.60 | 184.95 |
| GATA3 | 0.0020082 | 387 | GTEx | DepMap | Descartes | 1.66 | 297.09 |
| NNAT | 0.0019935 | 396 | GTEx | DepMap | Descartes | 0.61 | 245.78 |
VanGroningen Mesenchymal Genes
Mesenchymal marker genes from Supplementary Table 2 of Van Groningen et al. Nature Genetics 2017. These genes were identified by differential expression analysis of mesenchymal-like and adrenergic-like neuroblastoma cell lines.
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 13268.98
Median rank of genes in gene set: 15809.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| PPIB | 0.0022381 | 330 | GTEx | DepMap | Descartes | 0.66 | 258.76 |
| VIM | 0.0021859 | 341 | GTEx | DepMap | Descartes | 2.10 | 383.15 |
| PRDX4 | 0.0019206 | 414 | GTEx | DepMap | Descartes | 0.32 | 179.17 |
| CD63 | 0.0015854 | 567 | GTEx | DepMap | Descartes | 1.79 | 693.59 |
| POLR2L | 0.0015596 | 586 | GTEx | DepMap | Descartes | 0.40 | 215.04 |
| DDOST | 0.0014853 | 621 | GTEx | DepMap | Descartes | 0.40 | 98.74 |
| LAPTM4A | 0.0014533 | 636 | GTEx | DepMap | Descartes | 0.75 | 271.06 |
| PDIA6 | 0.0013814 | 693 | GTEx | DepMap | Descartes | 0.78 | 140.98 |
| MANF | 0.0013436 | 731 | GTEx | DepMap | Descartes | 0.29 | 94.67 |
| PRDX6 | 0.0013407 | 734 | GTEx | DepMap | Descartes | 0.57 | 164.07 |
| MYDGF | 0.0013157 | 755 | GTEx | DepMap | Descartes | 0.38 | NA |
| OSTC | 0.0012519 | 807 | GTEx | DepMap | Descartes | 0.45 | 214.61 |
| ROBO1 | 0.0011186 | 932 | GTEx | DepMap | Descartes | 10.33 | 792.53 |
| MYL12B | 0.0010901 | 969 | GTEx | DepMap | Descartes | 0.88 | 305.55 |
| EMILIN1 | 0.0010722 | 986 | GTEx | DepMap | Descartes | 0.39 | 54.77 |
| MYL12A | 0.0009971 | 1092 | GTEx | DepMap | Descartes | 0.34 | 126.30 |
| LHX8 | 0.0009369 | 1173 | GTEx | DepMap | Descartes | 0.04 | 6.63 |
| SOX9 | 0.0009099 | 1225 | GTEx | DepMap | Descartes | 0.12 | 17.65 |
| LGALS1 | 0.0008972 | 1243 | GTEx | DepMap | Descartes | 1.23 | 756.80 |
| ABRACL | 0.0008564 | 1314 | GTEx | DepMap | Descartes | 0.13 | NA |
| SSBP4 | 0.0007929 | 1434 | GTEx | DepMap | Descartes | 0.58 | 156.84 |
| SCRG1 | 0.0007852 | 1450 | GTEx | DepMap | Descartes | 0.12 | 14.56 |
| HLA-C | 0.0007423 | 1544 | GTEx | DepMap | Descartes | 0.31 | 65.13 |
| TRIL | 0.0007363 | 1555 | GTEx | DepMap | Descartes | 0.11 | 9.51 |
| GJA1 | 0.0007224 | 1579 | GTEx | DepMap | Descartes | 0.20 | 18.63 |
| MEST | 0.0007036 | 1617 | GTEx | DepMap | Descartes | 0.43 | 83.02 |
| TMED9 | 0.0006838 | 1671 | GTEx | DepMap | Descartes | 0.27 | 46.01 |
| SSR3 | 0.0006692 | 1701 | GTEx | DepMap | Descartes | 0.42 | 52.41 |
| KDM5B | 0.0006619 | 1726 | GTEx | DepMap | Descartes | 1.09 | 56.70 |
| TMBIM4 | 0.0005434 | 2062 | GTEx | DepMap | Descartes | 0.08 | 11.76 |
Descartes adrenocortical markers
Top 50 marker genes of adrenocortical cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/adrenocortical/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 13254.64
Median rank of genes in gene set: 16788
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| HSPE1 | 0.0032458 | 174 | GTEx | DepMap | Descartes | 0.86 | 594.01 |
| HSPD1 | 0.0027663 | 234 | GTEx | DepMap | Descartes | 2.09 | 437.23 |
| SGCZ | 0.0019905 | 397 | GTEx | DepMap | Descartes | 16.20 | 1299.50 |
| CYP21A2 | 0.0013136 | 758 | GTEx | DepMap | Descartes | 0.58 | 63.58 |
| FDXR | 0.0010164 | 1058 | GTEx | DepMap | Descartes | 0.16 | 22.27 |
| CYP17A1 | 0.0006888 | 1658 | GTEx | DepMap | Descartes | 0.46 | 55.33 |
| TM7SF2 | 0.0005040 | 2217 | GTEx | DepMap | Descartes | 0.21 | 41.56 |
| STAR | 0.0003674 | 2830 | GTEx | DepMap | Descartes | 0.67 | 48.97 |
| GSTA4 | 0.0002906 | 3278 | GTEx | DepMap | Descartes | 0.59 | 140.85 |
| DHCR7 | 0.0001331 | 4462 | GTEx | DepMap | Descartes | 0.12 | 18.50 |
| PEG3 | 0.0000508 | 5437 | GTEx | DepMap | Descartes | 0.01 | NA |
| INHA | 0.0000459 | 5520 | GTEx | DepMap | Descartes | 0.03 | 6.43 |
| CYB5B | -0.0000231 | 8062 | GTEx | DepMap | Descartes | 0.60 | 51.15 |
| JAKMIP2 | -0.0000863 | 10247 | GTEx | DepMap | Descartes | 1.40 | 64.35 |
| FDX1 | -0.0002426 | 13273 | GTEx | DepMap | Descartes | 1.84 | 151.99 |
| HMGCR | -0.0002580 | 13496 | GTEx | DepMap | Descartes | 0.58 | 50.42 |
| DHCR24 | -0.0003187 | 14260 | GTEx | DepMap | Descartes | 0.57 | 31.80 |
| MC2R | -0.0004303 | 15493 | GTEx | DepMap | Descartes | 0.14 | 9.37 |
| SLC16A9 | -0.0004346 | 15537 | GTEx | DepMap | Descartes | 0.45 | 41.36 |
| FREM2 | -0.0004410 | 15592 | GTEx | DepMap | Descartes | 0.04 | 0.71 |
| CYP11B1 | -0.0004874 | 16024 | GTEx | DepMap | Descartes | 1.05 | 66.64 |
| SLC1A2 | -0.0005521 | 16525 | GTEx | DepMap | Descartes | 0.79 | 29.73 |
| DNER | -0.0005833 | 16788 | GTEx | DepMap | Descartes | 0.70 | 70.43 |
| MSMO1 | -0.0006205 | 17057 | GTEx | DepMap | Descartes | 0.44 | 76.95 |
| FDPS | -0.0006344 | 17153 | GTEx | DepMap | Descartes | 0.91 | 177.88 |
| SLC2A14 | -0.0006419 | 17215 | GTEx | DepMap | Descartes | 0.05 | 3.69 |
| SCAP | -0.0006791 | 17475 | GTEx | DepMap | Descartes | 0.62 | 60.18 |
| APOC1 | -0.0007265 | 17757 | GTEx | DepMap | Descartes | 0.24 | 111.92 |
| NPC1 | -0.0007315 | 17778 | GTEx | DepMap | Descartes | 0.49 | 38.93 |
| FRMD5 | -0.0007472 | 17875 | GTEx | DepMap | Descartes | 2.91 | 241.29 |
Descartes chromaffin markers
Top 50 marker genes of chromaffin cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/chromaffin/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.27e-02
Mean rank of genes in gene set: 8271.07
Median rank of genes in gene set: 4288
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TUBB2B | 0.0060284 | 87 | GTEx | DepMap | Descartes | 3.63 | 1054.34 |
| STMN2 | 0.0033906 | 159 | GTEx | DepMap | Descartes | 8.67 | 2137.58 |
| BASP1 | 0.0033407 | 166 | GTEx | DepMap | Descartes | 2.71 | 809.68 |
| NPY | 0.0032694 | 171 | GTEx | DepMap | Descartes | 3.49 | 2692.04 |
| CCND1 | 0.0032207 | 181 | GTEx | DepMap | Descartes | 2.62 | 340.49 |
| MAB21L2 | 0.0031090 | 193 | GTEx | DepMap | Descartes | 0.30 | 81.67 |
| TUBA1A | 0.0029708 | 206 | GTEx | DepMap | Descartes | 8.21 | 2186.52 |
| GAL | 0.0025011 | 267 | GTEx | DepMap | Descartes | 2.19 | 1228.63 |
| ISL1 | 0.0021995 | 339 | GTEx | DepMap | Descartes | 0.94 | 221.35 |
| MAB21L1 | 0.0021786 | 343 | GTEx | DepMap | Descartes | 0.40 | 77.67 |
| STMN4 | 0.0021356 | 357 | GTEx | DepMap | Descartes | 1.07 | 232.45 |
| ELAVL2 | 0.0015620 | 580 | GTEx | DepMap | Descartes | 2.12 | 271.78 |
| EPHA6 | 0.0012351 | 831 | GTEx | DepMap | Descartes | 4.28 | 501.55 |
| EYA1 | 0.0009188 | 1206 | GTEx | DepMap | Descartes | 2.51 | 295.72 |
| KCNB2 | 0.0008401 | 1341 | GTEx | DepMap | Descartes | 3.15 | 422.06 |
| MLLT11 | 0.0007508 | 1523 | GTEx | DepMap | Descartes | 1.16 | 236.24 |
| RGMB | 0.0005230 | 2131 | GTEx | DepMap | Descartes | 0.49 | 48.68 |
| HS3ST5 | 0.0005131 | 2176 | GTEx | DepMap | Descartes | 0.82 | 82.80 |
| FAT3 | 0.0004965 | 2255 | GTEx | DepMap | Descartes | 1.06 | 30.01 |
| IL7 | 0.0003945 | 2684 | GTEx | DepMap | Descartes | 2.18 | 421.32 |
| CNTFR | 0.0002635 | 3428 | GTEx | DepMap | Descartes | 0.34 | 70.87 |
| SLC44A5 | 0.0001527 | 4288 | GTEx | DepMap | Descartes | 1.53 | 154.10 |
| TMEFF2 | 0.0001485 | 4331 | GTEx | DepMap | Descartes | 0.85 | 101.14 |
| HMX1 | 0.0000676 | 5225 | GTEx | DepMap | Descartes | 0.55 | 126.37 |
| PTCHD1 | -0.0000483 | 9085 | GTEx | DepMap | Descartes | 0.46 | 13.01 |
| GREM1 | -0.0000819 | 10134 | GTEx | DepMap | Descartes | 0.06 | 1.12 |
| SYNPO2 | -0.0002190 | 12943 | GTEx | DepMap | Descartes | 1.17 | 38.91 |
| EYA4 | -0.0003712 | 14863 | GTEx | DepMap | Descartes | 0.46 | 33.23 |
| CNKSR2 | -0.0003809 | 14966 | GTEx | DepMap | Descartes | 1.51 | 77.70 |
| RYR2 | -0.0003935 | 15093 | GTEx | DepMap | Descartes | 5.89 | 146.61 |
Descartes Vascular_endothelial markers
Top 50 marker genes of Vascular_endothelial cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/vascular_endothelial/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 9.74e-01
Mean rank of genes in gene set: 12036.43
Median rank of genes in gene set: 12679
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| RAMP2 | 0.0014511 | 638 | GTEx | DepMap | Descartes | 0.23 | 133.43 |
| HYAL2 | 0.0009651 | 1133 | GTEx | DepMap | Descartes | 0.22 | 25.47 |
| EFNB2 | 0.0003562 | 2899 | GTEx | DepMap | Descartes | 0.46 | 46.26 |
| ESM1 | 0.0002731 | 3375 | GTEx | DepMap | Descartes | 0.06 | 12.93 |
| SOX18 | 0.0002120 | 3797 | GTEx | DepMap | Descartes | 0.09 | 18.85 |
| ID1 | 0.0001867 | 4014 | GTEx | DepMap | Descartes | 0.23 | 67.67 |
| ARHGAP29 | 0.0001718 | 4129 | GTEx | DepMap | Descartes | 0.99 | 54.48 |
| CHRM3 | 0.0001405 | 4402 | GTEx | DepMap | Descartes | 1.55 | 81.31 |
| CLDN5 | 0.0000946 | 4875 | GTEx | DepMap | Descartes | 0.09 | 14.39 |
| IRX3 | 0.0000540 | 5389 | GTEx | DepMap | Descartes | 0.02 | 3.71 |
| TM4SF18 | 0.0000351 | 5689 | GTEx | DepMap | Descartes | 0.04 | 4.41 |
| TMEM88 | -0.0000451 | 8963 | GTEx | DepMap | Descartes | 0.04 | 18.14 |
| CYP26B1 | -0.0000634 | 9595 | GTEx | DepMap | Descartes | 0.02 | 2.44 |
| APLNR | -0.0000792 | 10056 | GTEx | DepMap | Descartes | 0.03 | 3.77 |
| ECSCR | -0.0000797 | 10070 | GTEx | DepMap | Descartes | 0.00 | 0.06 |
| NPR1 | -0.0000869 | 10261 | GTEx | DepMap | Descartes | 0.02 | 1.90 |
| PODXL | -0.0001359 | 11453 | GTEx | DepMap | Descartes | 0.27 | 18.40 |
| MMRN2 | -0.0001505 | 11744 | GTEx | DepMap | Descartes | 0.09 | 8.59 |
| DNASE1L3 | -0.0001668 | 12043 | GTEx | DepMap | Descartes | 0.06 | 11.75 |
| KDR | -0.0001878 | 12415 | GTEx | DepMap | Descartes | 0.07 | 5.19 |
| TIE1 | -0.0001955 | 12544 | GTEx | DepMap | Descartes | 0.06 | 5.72 |
| CRHBP | -0.0001987 | 12598 | GTEx | DepMap | Descartes | 0.02 | 3.10 |
| PLVAP | -0.0002086 | 12760 | GTEx | DepMap | Descartes | 0.16 | 28.73 |
| ROBO4 | -0.0002371 | 13206 | GTEx | DepMap | Descartes | 0.05 | 4.63 |
| RASIP1 | -0.0003175 | 14247 | GTEx | DepMap | Descartes | 0.06 | 7.06 |
| FLT4 | -0.0003191 | 14264 | GTEx | DepMap | Descartes | 0.03 | 2.02 |
| KANK3 | -0.0003643 | 14806 | GTEx | DepMap | Descartes | 0.05 | 6.65 |
| CDH5 | -0.0003944 | 15112 | GTEx | DepMap | Descartes | 0.08 | 7.21 |
| FCGR2B | -0.0003981 | 15144 | GTEx | DepMap | Descartes | 0.01 | 0.94 |
| BTNL9 | -0.0003988 | 15153 | GTEx | DepMap | Descartes | 0.06 | 5.98 |
Descartes stromal markers
Top 50 marker genes of stromal cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/stromal/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 13527.55
Median rank of genes in gene set: 15189
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| PCOLCE | 0.0016862 | 518 | GTEx | DepMap | Descartes | 0.61 | 217.22 |
| PCDH18 | 0.0003882 | 2720 | GTEx | DepMap | Descartes | 0.06 | 5.08 |
| CD248 | 0.0002476 | 3545 | GTEx | DepMap | Descartes | 0.05 | 10.40 |
| MGP | 0.0002353 | 3621 | GTEx | DepMap | Descartes | 1.49 | 334.95 |
| ZNF385D | 0.0001700 | 4148 | GTEx | DepMap | Descartes | 0.85 | 33.02 |
| C7 | 0.0001411 | 4393 | GTEx | DepMap | Descartes | 0.71 | 48.58 |
| POSTN | 0.0001219 | 4564 | GTEx | DepMap | Descartes | 0.57 | 66.78 |
| DKK2 | 0.0000918 | 4917 | GTEx | DepMap | Descartes | 0.15 | 19.00 |
| SFRP2 | -0.0000508 | 9164 | GTEx | DepMap | Descartes | 0.26 | 62.62 |
| LRRC17 | -0.0000514 | 9182 | GTEx | DepMap | Descartes | 0.09 | 19.22 |
| CLDN11 | -0.0000621 | 9552 | GTEx | DepMap | Descartes | 0.00 | 0.01 |
| SULT1E1 | -0.0000702 | 9806 | GTEx | DepMap | Descartes | 0.00 | 0.33 |
| OGN | -0.0001345 | 11420 | GTEx | DepMap | Descartes | 0.12 | 13.72 |
| RSPO3 | -0.0001553 | 11826 | GTEx | DepMap | Descartes | 0.03 | NA |
| HHIP | -0.0002344 | 13160 | GTEx | DepMap | Descartes | 0.33 | 15.34 |
| PRRX1 | -0.0002529 | 13426 | GTEx | DepMap | Descartes | 0.19 | 17.20 |
| SCARA5 | -0.0002632 | 13559 | GTEx | DepMap | Descartes | 0.01 | 0.71 |
| DCN | -0.0002647 | 13590 | GTEx | DepMap | Descartes | 0.45 | 23.50 |
| MXRA5 | -0.0002717 | 13681 | GTEx | DepMap | Descartes | 0.06 | 1.99 |
| PAMR1 | -0.0002946 | 13969 | GTEx | DepMap | Descartes | 0.19 | 19.44 |
| PDGFRA | -0.0003242 | 14329 | GTEx | DepMap | Descartes | 0.06 | 4.33 |
| ACTA2 | -0.0003297 | 14404 | GTEx | DepMap | Descartes | 0.36 | 99.62 |
| EDNRA | -0.0003766 | 14916 | GTEx | DepMap | Descartes | 0.15 | 13.27 |
| LAMC3 | -0.0003814 | 14969 | GTEx | DepMap | Descartes | 0.07 | 2.63 |
| COL6A3 | -0.0004026 | 15189 | GTEx | DepMap | Descartes | 0.55 | 22.07 |
| COL3A1 | -0.0004030 | 15196 | GTEx | DepMap | Descartes | 2.41 | 175.28 |
| FNDC1 | -0.0004550 | 15716 | GTEx | DepMap | Descartes | 0.06 | 3.65 |
| LUM | -0.0004707 | 15865 | GTEx | DepMap | Descartes | 0.44 | 65.96 |
| IGFBP3 | -0.0004831 | 15984 | GTEx | DepMap | Descartes | 0.17 | 24.62 |
| ABCC9 | -0.0004877 | 16028 | GTEx | DepMap | Descartes | 0.09 | 4.01 |
Descartes sympathoblasts markers
Top 50 marker genes of sympathoblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/sympathoblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 4.03e-01
Mean rank of genes in gene set: 10071.77
Median rank of genes in gene set: 12749
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CHGA | 0.0029321 | 211 | GTEx | DepMap | Descartes | 1.08 | 286.09 |
| CHGB | 0.0018495 | 453 | GTEx | DepMap | Descartes | 3.47 | 692.19 |
| GALNTL6 | 0.0015246 | 602 | GTEx | DepMap | Descartes | 2.12 | 263.86 |
| HTATSF1 | 0.0014319 | 654 | GTEx | DepMap | Descartes | 0.35 | 62.19 |
| PCSK1N | 0.0013866 | 687 | GTEx | DepMap | Descartes | 2.59 | 993.66 |
| KCTD16 | 0.0013853 | 689 | GTEx | DepMap | Descartes | 3.64 | 110.54 |
| INSM1 | 0.0013380 | 737 | GTEx | DepMap | Descartes | 0.18 | 33.13 |
| TENM1 | 0.0011495 | 900 | GTEx | DepMap | Descartes | 1.22 | NA |
| ROBO1 | 0.0011186 | 932 | GTEx | DepMap | Descartes | 10.33 | 792.53 |
| C1QL1 | 0.0010504 | 1012 | GTEx | DepMap | Descartes | 0.13 | 47.35 |
| PCSK2 | 0.0007568 | 1512 | GTEx | DepMap | Descartes | 0.75 | 62.98 |
| PENK | 0.0007564 | 1513 | GTEx | DepMap | Descartes | 0.05 | 24.68 |
| SPOCK3 | 0.0006324 | 1808 | GTEx | DepMap | Descartes | 1.11 | 179.10 |
| TBX20 | 0.0004516 | 2446 | GTEx | DepMap | Descartes | 0.31 | 87.48 |
| UNC80 | 0.0001963 | 3925 | GTEx | DepMap | Descartes | 1.53 | 47.83 |
| MGAT4C | 0.0001883 | 4001 | GTEx | DepMap | Descartes | 1.57 | 25.75 |
| PNMT | 0.0001737 | 4112 | GTEx | DepMap | Descartes | 0.01 | 5.87 |
| SORCS3 | 0.0001149 | 4635 | GTEx | DepMap | Descartes | 0.39 | 29.00 |
| SLC18A1 | 0.0000953 | 4863 | GTEx | DepMap | Descartes | 0.13 | 20.09 |
| LAMA3 | -0.0001516 | 11762 | GTEx | DepMap | Descartes | 0.48 | 21.98 |
| GRM7 | -0.0002027 | 12674 | GTEx | DepMap | Descartes | 2.04 | 236.55 |
| CNTNAP5 | -0.0002078 | 12749 | GTEx | DepMap | Descartes | 0.90 | 72.81 |
| CDH18 | -0.0002298 | 13097 | GTEx | DepMap | Descartes | 1.22 | 106.58 |
| GCH1 | -0.0003013 | 14043 | GTEx | DepMap | Descartes | 0.48 | 74.30 |
| NTNG1 | -0.0003025 | 14059 | GTEx | DepMap | Descartes | 2.18 | 223.11 |
| CNTN3 | -0.0003902 | 15061 | GTEx | DepMap | Descartes | 0.13 | 9.43 |
| FGF14 | -0.0004110 | 15291 | GTEx | DepMap | Descartes | 11.52 | 407.68 |
| ST18 | -0.0004977 | 16106 | GTEx | DepMap | Descartes | 0.07 | 4.43 |
| DGKK | -0.0005020 | 16134 | GTEx | DepMap | Descartes | 0.08 | 4.23 |
| SCG2 | -0.0005272 | 16357 | GTEx | DepMap | Descartes | 4.39 | 572.29 |
Descartes erythroblasts markers
Top 50 marker genes of erythroblasts cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/erythroblasts/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 13211.7
Median rank of genes in gene set: 13717.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| CAT | 0.0003132 | 3149 | GTEx | DepMap | Descartes | 0.30 | 58.08 |
| HBB | 0.0003079 | 3174 | GTEx | DepMap | Descartes | 0.25 | 204.62 |
| FECH | 0.0002623 | 3437 | GTEx | DepMap | Descartes | 0.21 | 12.12 |
| GYPC | 0.0002594 | 3458 | GTEx | DepMap | Descartes | 0.08 | 19.80 |
| HBA2 | 0.0001244 | 4543 | GTEx | DepMap | Descartes | 0.07 | 60.86 |
| HBA1 | 0.0000930 | 4900 | GTEx | DepMap | Descartes | 0.03 | 29.01 |
| RHD | 0.0000924 | 4909 | GTEx | DepMap | Descartes | 0.08 | 12.72 |
| HBG1 | 0.0000183 | 6049 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| HBG2 | 0.0000000 | 6896 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| GYPB | -0.0000264 | 8212 | GTEx | DepMap | Descartes | 0.02 | 14.14 |
| ALAS2 | -0.0001120 | 10922 | GTEx | DepMap | Descartes | 0.00 | 0.61 |
| HEMGN | -0.0001313 | 11352 | GTEx | DepMap | Descartes | 0.00 | 0.56 |
| HBZ | -0.0001324 | 11375 | GTEx | DepMap | Descartes | 0.00 | 2.58 |
| EPB42 | -0.0001335 | 11399 | GTEx | DepMap | Descartes | 0.00 | 0.27 |
| HBM | -0.0001370 | 11479 | GTEx | DepMap | Descartes | 0.00 | 3.38 |
| AHSP | -0.0001435 | 11604 | GTEx | DepMap | Descartes | 0.00 | 0.55 |
| GYPA | -0.0001450 | 11633 | GTEx | DepMap | Descartes | 0.01 | 1.34 |
| SELENBP1 | -0.0001471 | 11675 | GTEx | DepMap | Descartes | 0.01 | 1.27 |
| RHCE | -0.0001630 | 11973 | GTEx | DepMap | Descartes | 0.04 | 9.76 |
| CPOX | -0.0001941 | 12522 | GTEx | DepMap | Descartes | 0.03 | 4.61 |
| SPTB | -0.0002243 | 13022 | GTEx | DepMap | Descartes | 0.17 | 5.88 |
| RHAG | -0.0002360 | 13184 | GTEx | DepMap | Descartes | 0.00 | 0.72 |
| SPTA1 | -0.0002656 | 13605 | GTEx | DepMap | Descartes | 0.00 | 0.10 |
| TMCC2 | -0.0002833 | 13830 | GTEx | DepMap | Descartes | 0.09 | 9.30 |
| SLC4A1 | -0.0002932 | 13949 | GTEx | DepMap | Descartes | 0.01 | 0.58 |
| GYPE | -0.0003313 | 14427 | GTEx | DepMap | Descartes | 0.04 | 9.66 |
| CR1L | -0.0003795 | 14949 | GTEx | DepMap | Descartes | 0.02 | 5.26 |
| TSPAN5 | -0.0003829 | 14980 | GTEx | DepMap | Descartes | 1.63 | 193.13 |
| ABCB10 | -0.0004010 | 15168 | GTEx | DepMap | Descartes | 0.24 | 27.77 |
| XPO7 | -0.0004959 | 16092 | GTEx | DepMap | Descartes | 0.72 | 67.18 |
Descartes myeloid markers
Top 50 marker genes of myeloid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/myeloid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 14396.04
Median rank of genes in gene set: 16255
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| HLA-DPA1 | 0.0004545 | 2424 | GTEx | DepMap | Descartes | 0.21 | 19.01 |
| SPP1 | 0.0004106 | 2613 | GTEx | DepMap | Descartes | 1.06 | 261.88 |
| RBPJ | 0.0003094 | 3168 | GTEx | DepMap | Descartes | 1.52 | 127.84 |
| C1QC | 0.0001540 | 4279 | GTEx | DepMap | Descartes | 0.07 | 28.42 |
| C1QB | 0.0001507 | 4310 | GTEx | DepMap | Descartes | 0.11 | 45.05 |
| CTSC | 0.0000229 | 5937 | GTEx | DepMap | Descartes | 0.33 | 26.92 |
| C1QA | -0.0000032 | 7200 | GTEx | DepMap | Descartes | 0.11 | 48.04 |
| MS4A7 | -0.0000377 | 8682 | GTEx | DepMap | Descartes | 0.01 | 1.87 |
| CTSD | -0.0000491 | 9107 | GTEx | DepMap | Descartes | 0.00 | 0.01 |
| CD14 | -0.0000696 | 9794 | GTEx | DepMap | Descartes | 0.08 | 19.84 |
| CYBB | -0.0000768 | 9993 | GTEx | DepMap | Descartes | 0.01 | 0.62 |
| FGL2 | -0.0001074 | 10812 | GTEx | DepMap | Descartes | 0.06 | 6.06 |
| HLA-DRA | -0.0001196 | 11095 | GTEx | DepMap | Descartes | 0.29 | 105.29 |
| MPEG1 | -0.0001301 | 11325 | GTEx | DepMap | Descartes | 0.03 | 3.39 |
| RNASE1 | -0.0001596 | 11906 | GTEx | DepMap | Descartes | 0.07 | 28.38 |
| CD74 | -0.0001765 | 12214 | GTEx | DepMap | Descartes | 0.59 | 90.11 |
| LGMN | -0.0002527 | 13421 | GTEx | DepMap | Descartes | 0.26 | 54.74 |
| VSIG4 | -0.0002858 | 13871 | GTEx | DepMap | Descartes | 0.01 | 2.14 |
| IFNGR1 | -0.0004127 | 15305 | GTEx | DepMap | Descartes | 0.25 | 42.99 |
| MS4A6A | -0.0004217 | 15412 | GTEx | DepMap | Descartes | 0.09 | 16.74 |
| CPVL | -0.0004276 | 15472 | GTEx | DepMap | Descartes | 0.22 | 39.94 |
| FGD2 | -0.0004329 | 15511 | GTEx | DepMap | Descartes | 0.03 | 2.30 |
| MS4A4A | -0.0004724 | 15881 | GTEx | DepMap | Descartes | 0.06 | 15.23 |
| HLA-DRB1 | -0.0005101 | 16212 | GTEx | DepMap | Descartes | 0.06 | 20.12 |
| TGFBI | -0.0005150 | 16255 | GTEx | DepMap | Descartes | 0.18 | 14.39 |
| HRH1 | -0.0005502 | 16511 | GTEx | DepMap | Descartes | 0.23 | 14.68 |
| CSF1R | -0.0005650 | 16631 | GTEx | DepMap | Descartes | 0.06 | 6.23 |
| CD163 | -0.0005784 | 16743 | GTEx | DepMap | Descartes | 0.04 | 3.32 |
| SLC1A3 | -0.0006716 | 17418 | GTEx | DepMap | Descartes | 0.20 | 18.13 |
| MS4A4E | -0.0006889 | 17531 | GTEx | DepMap | Descartes | 0.05 | 8.66 |
Descartes Schwann markers
Top 50 marker genes of Schwann cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/schwann/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 14344.71
Median rank of genes in gene set: 17142
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| MARCKS | 0.0041567 | 125 | GTEx | DepMap | Descartes | 2.09 | 279.84 |
| VIM | 0.0021859 | 341 | GTEx | DepMap | Descartes | 2.10 | 383.15 |
| GFRA3 | 0.0012971 | 768 | GTEx | DepMap | Descartes | 0.18 | 51.19 |
| SFRP1 | 0.0008329 | 1356 | GTEx | DepMap | Descartes | 0.49 | 45.63 |
| SLC35F1 | 0.0005241 | 2124 | GTEx | DepMap | Descartes | 1.22 | 121.65 |
| MDGA2 | 0.0003438 | 2976 | GTEx | DepMap | Descartes | 0.96 | 46.33 |
| PTN | 0.0001540 | 4278 | GTEx | DepMap | Descartes | 0.48 | 123.83 |
| SOX5 | 0.0001478 | 4336 | GTEx | DepMap | Descartes | 3.41 | 238.68 |
| KCTD12 | 0.0001464 | 4350 | GTEx | DepMap | Descartes | 0.22 | 15.18 |
| LRRTM4 | 0.0000322 | 5758 | GTEx | DepMap | Descartes | 2.98 | 303.20 |
| PAG1 | -0.0001223 | 11153 | GTEx | DepMap | Descartes | 0.98 | 42.42 |
| HMGA2 | -0.0001615 | 11943 | GTEx | DepMap | Descartes | 0.04 | 2.10 |
| ADAMTS5 | -0.0001960 | 12553 | GTEx | DepMap | Descartes | 0.08 | 3.15 |
| FIGN | -0.0002282 | 13073 | GTEx | DepMap | Descartes | 0.64 | 31.61 |
| IL1RAPL1 | -0.0002646 | 13587 | GTEx | DepMap | Descartes | 1.19 | 118.36 |
| NRXN1 | -0.0002709 | 13675 | GTEx | DepMap | Descartes | 9.64 | 452.83 |
| MPZ | -0.0003702 | 14852 | GTEx | DepMap | Descartes | 0.05 | 7.33 |
| SOX10 | -0.0003920 | 15077 | GTEx | DepMap | Descartes | 0.05 | 6.25 |
| ERBB3 | -0.0004299 | 15489 | GTEx | DepMap | Descartes | 0.02 | 1.30 |
| VCAN | -0.0004436 | 15608 | GTEx | DepMap | Descartes | 1.38 | 50.23 |
| OLFML2A | -0.0005367 | 16425 | GTEx | DepMap | Descartes | 0.06 | 3.07 |
| PLP1 | -0.0006030 | 16928 | GTEx | DepMap | Descartes | 0.05 | 4.75 |
| XKR4 | -0.0006202 | 17055 | GTEx | DepMap | Descartes | 0.63 | 11.12 |
| PTPRZ1 | -0.0006290 | 17114 | GTEx | DepMap | Descartes | 0.09 | 3.53 |
| LAMB1 | -0.0006330 | 17142 | GTEx | DepMap | Descartes | 1.04 | 82.64 |
| SCN7A | -0.0006415 | 17211 | GTEx | DepMap | Descartes | 0.58 | 24.62 |
| IL1RAPL2 | -0.0006687 | 17395 | GTEx | DepMap | Descartes | 0.52 | 75.95 |
| COL25A1 | -0.0006720 | 17424 | GTEx | DepMap | Descartes | 0.17 | 9.79 |
| EDNRB | -0.0006771 | 17457 | GTEx | DepMap | Descartes | 0.07 | 5.95 |
| SORCS1 | -0.0006792 | 17476 | GTEx | DepMap | Descartes | 1.76 | 87.94 |
Descartes Megakaryocytes markers
Top 50 marker genes of Megakaryocytes cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/megakaryocytes/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 14982.8
Median rank of genes in gene set: 16305
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| ACTB | 0.0044160 | 115 | GTEx | DepMap | Descartes | 5.51 | 1380.08 |
| TMSB4X | 0.0038437 | 138 | GTEx | DepMap | Descartes | 4.48 | 1383.48 |
| PRKAR2B | 0.0000258 | 5877 | GTEx | DepMap | Descartes | 1.21 | 125.09 |
| GP9 | 0.0000147 | 6124 | GTEx | DepMap | Descartes | 0.00 | 0.04 |
| TUBB1 | 0.0000147 | 6127 | GTEx | DepMap | Descartes | 0.01 | 0.91 |
| PPBP | -0.0000344 | 8543 | GTEx | DepMap | Descartes | 0.00 | 0.55 |
| SPN | -0.0000633 | 9589 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| PF4 | -0.0000817 | 10128 | GTEx | DepMap | Descartes | 0.00 | 0.54 |
| DOK6 | -0.0000972 | 10556 | GTEx | DepMap | Descartes | 1.15 | 55.87 |
| GP1BA | -0.0001002 | 10630 | GTEx | DepMap | Descartes | 0.01 | 1.41 |
| ITGB3 | -0.0001009 | 10646 | GTEx | DepMap | Descartes | 0.00 | 0.01 |
| MED12L | -0.0002336 | 13151 | GTEx | DepMap | Descartes | 0.67 | 27.87 |
| ANGPT1 | -0.0003047 | 14099 | GTEx | DepMap | Descartes | 0.17 | 15.65 |
| RAB27B | -0.0003433 | 14560 | GTEx | DepMap | Descartes | 0.27 | 15.03 |
| P2RX1 | -0.0003675 | 14827 | GTEx | DepMap | Descartes | 0.01 | 0.94 |
| MMRN1 | -0.0003784 | 14934 | GTEx | DepMap | Descartes | 0.08 | 5.51 |
| BIN2 | -0.0003947 | 15115 | GTEx | DepMap | Descartes | 0.03 | 6.60 |
| CD84 | -0.0004274 | 15470 | GTEx | DepMap | Descartes | 0.03 | 2.17 |
| STOM | -0.0004610 | 15775 | GTEx | DepMap | Descartes | 0.34 | 41.97 |
| PDE3A | -0.0004670 | 15818 | GTEx | DepMap | Descartes | 2.11 | 109.96 |
| FLNA | -0.0004764 | 15911 | GTEx | DepMap | Descartes | 0.40 | 20.71 |
| PLEK | -0.0004977 | 16105 | GTEx | DepMap | Descartes | 0.03 | 4.47 |
| PSTPIP2 | -0.0005064 | 16180 | GTEx | DepMap | Descartes | 0.07 | 9.54 |
| ITGA2B | -0.0005151 | 16256 | GTEx | DepMap | Descartes | 0.07 | 7.84 |
| FERMT3 | -0.0005206 | 16305 | GTEx | DepMap | Descartes | 0.04 | 7.44 |
| TGFB1 | -0.0005318 | 16389 | GTEx | DepMap | Descartes | 0.28 | 41.60 |
| SLC2A3 | -0.0005637 | 16614 | GTEx | DepMap | Descartes | 0.43 | 35.87 |
| STON2 | -0.0005686 | 16658 | GTEx | DepMap | Descartes | 0.20 | 19.63 |
| MYLK | -0.0005915 | 16845 | GTEx | DepMap | Descartes | 0.30 | 10.89 |
| TLN1 | -0.0005958 | 16878 | GTEx | DepMap | Descartes | 0.34 | 15.70 |
Descartes Lyphoid markers
Top 50 marker genes of Lyphoid cells in the Decartes fetal adrenal single cell map (https://atlas.brotmanbaty.org/bbi/human-gene-expression-during-development/cell/lymphoid/in/adrenal)
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.00e+00
Mean rank of genes in gene set: 15163.72
Median rank of genes in gene set: 17490
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TMSB10 | 0.0080109 | 78 | GTEx | DepMap | Descartes | 8.53 | 10924.99 |
| HLA-C | 0.0007423 | 1544 | GTEx | DepMap | Descartes | 0.31 | 65.13 |
| FOXP1 | 0.0005002 | 2237 | GTEx | DepMap | Descartes | 0.30 | 16.87 |
| HLA-A | 0.0001740 | 4108 | GTEx | DepMap | Descartes | 0.65 | 45.89 |
| TOX | 0.0001682 | 4164 | GTEx | DepMap | Descartes | 2.48 | 333.70 |
| GNG2 | 0.0001154 | 4628 | GTEx | DepMap | Descartes | 1.16 | 128.45 |
| SCML4 | -0.0000110 | 7531 | GTEx | DepMap | Descartes | 0.50 | 56.08 |
| NKG7 | -0.0000870 | 10262 | GTEx | DepMap | Descartes | 0.01 | 4.75 |
| EVL | -0.0000995 | 10615 | GTEx | DepMap | Descartes | 2.22 | 287.43 |
| ARHGDIB | -0.0001086 | 10842 | GTEx | DepMap | Descartes | 0.08 | 35.32 |
| BCL2 | -0.0001239 | 11206 | GTEx | DepMap | Descartes | 4.86 | 281.69 |
| LEF1 | -0.0001404 | 11540 | GTEx | DepMap | Descartes | 0.33 | 36.36 |
| HLA-B | -0.0002009 | 12652 | GTEx | DepMap | Descartes | 0.35 | 94.08 |
| SORL1 | -0.0002165 | 12898 | GTEx | DepMap | Descartes | 0.58 | 24.61 |
| PDE3B | -0.0003356 | 14473 | GTEx | DepMap | Descartes | 1.54 | 116.88 |
| CD44 | -0.0003483 | 14613 | GTEx | DepMap | Descartes | 1.50 | 124.62 |
| B2M | -0.0003670 | 14821 | GTEx | DepMap | Descartes | 3.71 | 627.56 |
| ETS1 | -0.0003788 | 14938 | GTEx | DepMap | Descartes | 0.34 | 30.99 |
| CCL5 | -0.0003898 | 15054 | GTEx | DepMap | Descartes | 0.02 | 8.72 |
| ITPKB | -0.0005448 | 16478 | GTEx | DepMap | Descartes | 0.16 | 11.41 |
| LCP1 | -0.0006285 | 17111 | GTEx | DepMap | Descartes | 0.12 | 13.88 |
| RCSD1 | -0.0006302 | 17121 | GTEx | DepMap | Descartes | 0.11 | 10.28 |
| BACH2 | -0.0006684 | 17392 | GTEx | DepMap | Descartes | 2.00 | 94.08 |
| IFI16 | -0.0006814 | 17490 | GTEx | DepMap | Descartes | 0.35 | 33.65 |
| IKZF1 | -0.0008223 | 18265 | GTEx | DepMap | Descartes | 0.14 | 12.30 |
| SKAP1 | -0.0008410 | 18367 | GTEx | DepMap | Descartes | 0.24 | 75.28 |
| CCND3 | -0.0008512 | 18433 | GTEx | DepMap | Descartes | 0.51 | 91.48 |
| STK39 | -0.0008545 | 18443 | GTEx | DepMap | Descartes | 1.43 | 194.07 |
| SAMD3 | -0.0009332 | 18775 | GTEx | DepMap | Descartes | 0.50 | 73.12 |
| MCTP2 | -0.0009433 | 18812 | GTEx | DepMap | Descartes | 0.22 | 10.64 |
Embryonic Pausing Signatures Up
Embryonic pausing signature upregulated genes from Supplementary Table 6 of Rehman et al. Cell 2021. These genes are upregulated in paused ESCs and diapaused embryos.
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.65e-01
Mean rank of genes in gene set: 8759.43
Median rank of genes in gene set: 7236.5
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| DPP7 | 0.0009047 | 1230 | GTEx | DepMap | Descartes | 0.44 | 121.38 |
| CCNG2 | 0.0005631 | 2001 | GTEx | DepMap | Descartes | 0.34 | 31.50 |
| APOE | 0.0002212 | 3725 | GTEx | DepMap | Descartes | 1.10 | 355.78 |
| PDCD4 | 0.0001318 | 4479 | GTEx | DepMap | Descartes | 0.59 | 75.23 |
| HEXA | 0.0001104 | 4687 | GTEx | DepMap | Descartes | 0.04 | 2.41 |
| ALDH6A1 | 0.0000435 | 5556 | GTEx | DepMap | Descartes | 0.10 | 8.59 |
| LY6G6E | 0.0000000 | 6989 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SPRY1 | -0.0000101 | 7484 | GTEx | DepMap | Descartes | 0.26 | 37.89 |
| RENBP | -0.0000422 | 8849 | GTEx | DepMap | Descartes | 0.07 | 20.31 |
| CTSL | -0.0000814 | 10120 | GTEx | DepMap | Descartes | 0.38 | NA |
| HEXB | -0.0003026 | 14061 | GTEx | DepMap | Descartes | 0.22 | 45.47 |
| ACSS1 | -0.0005811 | 16761 | GTEx | DepMap | Descartes | 0.05 | 3.59 |
| YPEL2 | -0.0006298 | 17119 | GTEx | DepMap | Descartes | 0.55 | 37.97 |
| AUH | -0.0011716 | 19571 | GTEx | DepMap | Descartes | 0.98 | 250.97 |
Embryonic Pausing Signatures Down
Embryonic pausing signature downregulated genes from Supplementary Table 6 of Rehman et al. Cell 2021. These genes are downregulated in paused ESCs and diapaused embryos.
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.33e-26
Mean rank of genes in gene set: 4155.57
Median rank of genes in gene set: 1744
Rank on gene expression program of top 30 genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Descartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| RPL41 | 0.0122859 | 40 | GTEx | DepMap | Descartes | 6.27 | 5388.55 |
| RANBP1 | 0.0032601 | 172 | GTEx | DepMap | Descartes | 0.74 | 211.20 |
| HSPE1 | 0.0032458 | 174 | GTEx | DepMap | Descartes | 0.86 | 594.01 |
| PPA1 | 0.0031751 | 184 | GTEx | DepMap | Descartes | 1.34 | 197.34 |
| SNRPE | 0.0031569 | 189 | GTEx | DepMap | Descartes | 0.61 | 229.30 |
| SNRPD2 | 0.0031318 | 190 | GTEx | DepMap | Descartes | 0.65 | 419.38 |
| SLC25A5 | 0.0031148 | 192 | GTEx | DepMap | Descartes | 0.40 | 153.96 |
| RAN | 0.0030176 | 202 | GTEx | DepMap | Descartes | 1.43 | 327.24 |
| HMGA1 | 0.0028512 | 222 | GTEx | DepMap | Descartes | 0.42 | 122.03 |
| MDH2 | 0.0026951 | 246 | GTEx | DepMap | Descartes | 0.65 | 161.41 |
| POLR2I | 0.0024567 | 282 | GTEx | DepMap | Descartes | 0.40 | 213.81 |
| TMEM258 | 0.0021373 | 356 | GTEx | DepMap | Descartes | 0.72 | NA |
| PHB | 0.0020733 | 378 | GTEx | DepMap | Descartes | 0.67 | 173.18 |
| RPS27L | 0.0020730 | 379 | GTEx | DepMap | Descartes | 0.53 | 42.58 |
| PA2G4 | 0.0019946 | 395 | GTEx | DepMap | Descartes | 0.43 | 105.72 |
| PEBP1 | 0.0019548 | 402 | GTEx | DepMap | Descartes | 2.56 | 756.75 |
| UQCR10 | 0.0019387 | 408 | GTEx | DepMap | Descartes | 0.85 | 480.63 |
| UQCRQ | 0.0019377 | 409 | GTEx | DepMap | Descartes | 0.70 | 233.95 |
| PDCD5 | 0.0019001 | 421 | GTEx | DepMap | Descartes | 0.55 | 148.63 |
| TMEM97 | 0.0018761 | 437 | GTEx | DepMap | Descartes | 0.25 | 55.96 |
| PRDX2 | 0.0017685 | 485 | GTEx | DepMap | Descartes | 0.26 | 75.81 |
| RPS19BP1 | 0.0017134 | 505 | GTEx | DepMap | Descartes | 0.40 | 69.28 |
| NDUFS8 | 0.0016119 | 556 | GTEx | DepMap | Descartes | 0.49 | 77.58 |
| DCTPP1 | 0.0015876 | 566 | GTEx | DepMap | Descartes | 0.16 | 74.77 |
| NDUFC1 | 0.0015603 | 583 | GTEx | DepMap | Descartes | 0.41 | 152.20 |
| POLR2L | 0.0015596 | 586 | GTEx | DepMap | Descartes | 0.40 | 215.04 |
| ELOF1 | 0.0014486 | 642 | GTEx | DepMap | Descartes | 0.16 | 38.49 |
| CCDC28B | 0.0014270 | 655 | GTEx | DepMap | Descartes | 0.15 | 63.35 |
| SAC3D1 | 0.0014246 | 658 | GTEx | DepMap | Descartes | 0.22 | 67.29 |
| PHGDH | 0.0014161 | 664 | GTEx | DepMap | Descartes | 0.22 | 41.74 |
Below shows the ranks on this GEP for immune marker gene lists curated by CellTypist (https://www.celltypist.org/encyclopedia).
High ranks indicate this gene is a driver of this GEP.
These curated gene list are ranked by P-value (on this GEP) of their constituent genes.
The Mean Count column shows the mean read count in cells scoring highly (H > 50) on this gene expression program.
Cycling cells: Cycling T cells (curated markers)
proliferating T lymphocytes:
Wilcoxon ranksum test P-value for gene set overrepresentation: 4.41e-03
Mean rank of genes in gene set: 1307
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| TOP2A | 0.0010713 | 989 | GTEx | DepMap | Descartes | 0.79 | 88.56 |
| CD3D | 0.0008723 | 1287 | GTEx | DepMap | Descartes | 0.03 | 23.95 |
| MKI67 | 0.0006942 | 1645 | GTEx | DepMap | Descartes | 0.28 | 14.02 |
T cells: CD8a/b(entry) (model markers)
T lymphocytes with CD8 alpha/beta heterodimers in the late double-positive (entry) stage during T cell development:
Wilcoxon ranksum test P-value for gene set overrepresentation: 1.03e-02
Mean rank of genes in gene set: 5705.56
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| RPL10 | 0.0153615 | 4 | GTEx | DepMap | Descartes | 10.55 | 2725.06 |
| RPS27 | 0.0144188 | 10 | GTEx | DepMap | Descartes | 6.70 | 6659.67 |
| TMSB10 | 0.0080109 | 78 | GTEx | DepMap | Descartes | 8.53 | 10924.99 |
| FTL | 0.0009297 | 1188 | GTEx | DepMap | Descartes | 4.37 | 2343.26 |
| MT-ND2 | 0.0002003 | 3902 | GTEx | DepMap | Descartes | 8.03 | 3170.55 |
| ITM2A | 0.0001411 | 4394 | GTEx | DepMap | Descartes | 0.04 | 10.33 |
| SATB1 | -0.0001020 | 10679 | GTEx | DepMap | Descartes | 0.01 | 0.47 |
| CD44 | -0.0003483 | 14613 | GTEx | DepMap | Descartes | 1.50 | 124.62 |
| CD1E | -0.0005451 | 16482 | GTEx | DepMap | Descartes | 0.01 | 0.79 |
HSC/MPP: Early lymphoid/T lymphoid (model markers)
early lymphoid/T lymphocytes with lymphocyte potential in the fetal liver before T cells emerged from the thymus:
Wilcoxon ranksum test P-value for gene set overrepresentation: 2.64e-02
Mean rank of genes in gene set: 6235.38
Rank on gene expression program of genes in gene set:
| Genes | Weight | Rank | GTEx | DepMap | Decartes | Mean.Counts | Mean.TPM |
|---|---|---|---|---|---|---|---|
| RACK1 | 0.0123656 | 39 | GTEx | DepMap | Descartes | 4.71 | NA |
| ATP5F1E | 0.0048155 | 104 | GTEx | DepMap | Descartes | 2.90 | NA |
| HBA2 | 0.0001244 | 4543 | GTEx | DepMap | Descartes | 0.07 | 60.86 |
| HBA1 | 0.0000930 | 4900 | GTEx | DepMap | Descartes | 0.03 | 29.01 |
| HBG2 | 0.0000000 | 6896 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| IGHV5-78 | -0.0000245 | 8126 | GTEx | DepMap | Descartes | 0.00 | 0.00 |
| SOST | -0.0000511 | 9169 | GTEx | DepMap | Descartes | 0.00 | 0.05 |
| ST18 | -0.0004977 | 16106 | GTEx | DepMap | Descartes | 0.07 | 4.43 |